Ecological Processes of Bacterial and Fungal Communities Associated with Typha orientalis Roots in Wetlands Were Distinct during Plant Development
- PMID: 36688664
- PMCID: PMC9927475
- DOI: 10.1128/spectrum.05051-22
Ecological Processes of Bacterial and Fungal Communities Associated with Typha orientalis Roots in Wetlands Were Distinct during Plant Development
Abstract
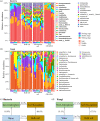
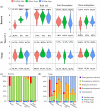
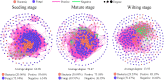
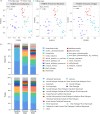
Root-associated microbiomes are essential for the ecological function of the root system. However, their assembly mechanisms in wetland are poorly understood. In this study, we explored and compared the ecological processes of bacterial and fungal communities in water, bulk soil, rhizosphere soil, and root endosphere niches for 3 developmental stages of Typha orientalis at different wetland sites, and assessed the potential functions of root endosphere microbiomes with function prediction. Our findings suggest that the microbial diversity, composition, and interaction networks along the water-soil-plant continuum are shaped predominantly by compartment niche and developmental stage, rather than by wetland site. Source tracking analysis indicated that T. orientalis' root endosphere is derived primarily from the rhizosphere soil (bacteria 39.9%, fungi 27.3%) and water (bacteria 18.9%, fungi 19.1%) niches. In addition, we found that the assembly of bacterial communities is driven primarily by deterministic processes and fungal communities by stochastic processes. The interaction network among microbes varies at different developmental stages of T. orientalis, and is accompanied by changes in microbial keystone taxa. The functional prediction data supports the distribution pattern of the bacterial and fungal microbiomes, which have different ecological roles at different plant developmental stages, where more beneficial bacterial taxa are observed in the root endosphere in the early stages, but more saprophytic fungi in the late stages. Our findings provide empirical evidence for the assembly, sources, interactions, and potential functions of wetland plant root microbial communities and have significant implications for the future applications of plant microbiomes in the wetland ecosystem. IMPORTANCE Our findings provide empirical evidence for the assembly, sources, interactions, and potential functions of wetland plant root microbial communities, and have significant implications for the future applications of plant microbiomes in the wetland ecosystem.
Keywords: Typha orientalis; developmental stage; ecological process; root endosphere; root-associated microbiome; wetland.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Divergent biotic and abiotic filtering of root endosphere and rhizosphere soil fungal communities along ecological gradients.FEMS Microbiol Ecol. 2020 Jul 1;96(7):fiaa124. doi: 10.1093/femsec/fiaa124. FEMS Microbiol Ecol. 2020. PMID: 32562419
-
Community Assembly of Fungi and Bacteria along Soil-Plant Continuum Differs in a Zoige Wetland.Microbiol Spectr. 2022 Oct 26;10(5):e0226022. doi: 10.1128/spectrum.02260-22. Epub 2022 Sep 22. Microbiol Spectr. 2022. PMID: 36135597 Free PMC article.
-
Plant domestication shapes rhizosphere microbiome assembly and metabolic functions.Microbiome. 2023 Mar 31;11(1):70. doi: 10.1186/s40168-023-01513-1. Microbiome. 2023. PMID: 37004105 Free PMC article.
-
Plant-microbe interactions through a lens: tales from the mycorrhizosphere.Ann Bot. 2024 Apr 10;133(3):399-412. doi: 10.1093/aob/mcad191. Ann Bot. 2024. PMID: 38085925 Free PMC article. Review.
-
Rhizosphere microbiome assembly, drivers and functions in perennial ligneous plant health.Microbiol Res. 2024 Oct;287:127860. doi: 10.1016/j.micres.2024.127860. Epub 2024 Jul 29. Microbiol Res. 2024. PMID: 39089083 Review.
Cited by
-
Unraveling the interplay between root exudates, microbiota, and rhizosheath formation in pearl millet.Microbiome. 2024 Jan 3;12(1):1. doi: 10.1186/s40168-023-01727-3. Microbiome. 2024. PMID: 38167150 Free PMC article.
-
Microbial degradation of naphthenic acids using constructed wetland treatment systems: metabolic and genomic insights for improved bioremediation of process-affected water.FEMS Microbiol Ecol. 2023 Nov 13;99(12):fiad153. doi: 10.1093/femsec/fiad153. FEMS Microbiol Ecol. 2023. PMID: 38012121 Free PMC article.
-
Characterization of endophytic bacteriome diversity and associated beneficial bacteria inhabiting a macrophyte Eichhornia crassipes.Front Plant Sci. 2023 Jun 19;14:1176648. doi: 10.3389/fpls.2023.1176648. eCollection 2023. Front Plant Sci. 2023. PMID: 37404529 Free PMC article.
-
Rhizosphere microbial ecological characteristics of strawberry root rot.Front Microbiol. 2023 Nov 16;14:1286740. doi: 10.3389/fmicb.2023.1286740. eCollection 2023. Front Microbiol. 2023. PMID: 38033596 Free PMC article.
-
Beyond correlation: Understanding the causal link between microbiome and plant health.Heliyon. 2024 Nov 19;10(23):e40517. doi: 10.1016/j.heliyon.2024.e40517. eCollection 2024 Dec 15. Heliyon. 2024. PMID: 39669148 Free PMC article. Review.
References
-
- Hu L, Robert C, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, van der Heijden M, Schlaeppi K, Erb M. 2018. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9:2738. doi:10.1038/s41467-018-05122-7. - DOI - PMC - PubMed
-
- Bulgarelli D, Rott M, Schlaeppi K, Ver LVTE, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi:10.1038/nature11336. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

