Modelling ribosome kinetics and translational control on dynamic mRNA
- PMID: 36689464
- PMCID: PMC9894550
- DOI: 10.1371/journal.pcbi.1010870
Modelling ribosome kinetics and translational control on dynamic mRNA
Abstract
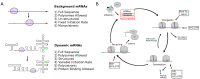
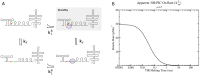
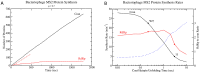
The control of protein synthesis and the overall levels of various proteins in the cell is critical for achieving homoeostasis. Regulation of protein levels can occur at the transcriptional level, where the total number of messenger RNAs in the overall transcriptome are controlled, or at the translational level, where interactions of proteins and ribosomes with the messenger RNA determine protein translational efficiency. Although transcriptional control of mRNA levels is the most commonly used regulatory control mechanism in cells, positive-sense single-stranded RNA viruses often utilise translational control mechanisms to regulate their proteins in the host cell. Here I detail a computational method for stochastically simulating protein synthesis on a dynamic messenger RNA using the Gillespie algorithm, where the mRNA is allowed to co-translationally fold in response to ribosome movement. Applying the model to the test case of the bacteriophage MS2 virus, I show that the models ability to accurately reproduce experimental measurements of coat protein production and translational repression of the viral RNA dependant RNA polymerase at high coat protein concentrations. The computational techniques reported here open up the potential to examine the infection dynamics of a ssRNA virus in a host cell at the level of the genomic RNA, as well as examine general translation control mechanisms present in polycistronic mRNAs.
Copyright: © 2023 Eric C. Dykeman. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The Author declares no competing interests.
Figures



References
-
- Calendar R. The Bacteriophages: Volume 1. Springer Science & Business Media; 2012.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

