Tissue-Resident Memory T Cells in Pancreatic Ductal Adenocarcinoma Coexpress PD-1 and TIGIT and Functional Inhibition Is Reversible by Dual Antibody Blockade
- PMID: 36689623
- PMCID: PMC10068448
- DOI: 10.1158/2326-6066.CIR-22-0121
Tissue-Resident Memory T Cells in Pancreatic Ductal Adenocarcinoma Coexpress PD-1 and TIGIT and Functional Inhibition Is Reversible by Dual Antibody Blockade
Abstract
Pancreatic ductal adenocarcinoma (PDAC) has a poor clinical outlook. Responses to immune checkpoint blockade are suboptimal and a much more detailed understanding of the tumor immune microenvironment is needed if this situation is to be improved. Here, we characterized tumor-infiltrating T-cell populations in patients with PDAC using cytometry by time of flight (CyTOF) and single-cell RNA sequencing. T cells were the predominant immune cell subset observed within tumors. Over 30% of CD4+ T cells expressed a CCR6+CD161+ Th17 phenotype and 17% displayed an activated regulatory T-cell profile. Large populations of CD8+ tissue-resident memory (TRM) T cells were also present and expressed high levels of programmed cell death protein 1 (PD-1) and TIGIT. A population of putative tumor-reactive CD103+CD39+ T cells was also observed within the CD8+ tumor-infiltrating lymphocytes population. The expression of PD-1 ligands was limited largely to hemopoietic cells whilst TIGIT ligands were expressed widely within the tumor microenvironment. Programmed death-ligand 1 and CD155 were expressed within the T-cell area of ectopic lymphoid structures and colocalized with PD-1+TIGIT+ CD8+ T cells. Combinatorial anti-PD-1 and TIGIT blockade enhanced IFNγ secretion and proliferation of T cells in the presence of PD-1 and TIGIT ligands. As such, we showed that the PDAC microenvironment is characterized by the presence of substantial populations of TRM cells with an exhausted PD-1+TIGIT+ phenotype where dual checkpoint receptor blockade represents a promising avenue for future immunotherapy.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures

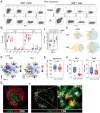
![Figure 2. Characterization of CD4+ T-cell populations within the PDAC tumor microenvironment. A, Dotplot of the top markers expressed in each CD4+ T-cell cluster identified via Louvain clustering of scRNA-seq data from CD4+ T cells from 3 PDAC tumor tissue samples (i). Where identifiable in the data, clusters are annotated with known CD4+ T-cell phenotypes. UMAP embedding of CD4+ T cells overlaid with Louvain cluster labels (ii). B, A 35-parameter CyTOF analysis of CD45+ cells from PDAC patient PBMC and TIL (n = 10). t-SNE plots shows Phenograph-clustered CD4+ T-cell populations in PBMC and TIL. C, Stacked bar graph showing the proportion of Naïve, EM, CM, and TEMRA subsets in CD4+ T cells generated from the data in (B). D, Bar graph comparing the proportion of each annotated CD4+ EM subset (TEM1-5) in PBMC vs. TIL, generated using the data in (B). E, Differential expression analysis distinguishing CD4+ Th17 from other non-Treg CD4+ T-cells in scRNA-seq data, first presented in Fig. 1. Selected genes are labelled, and colored points indicate genes that are differentially expressed [BH adjusted P < 0.01 and absolute (average logFC) > 0.5]. F, Quantification of CD4+ Th17 based on dual expression of CCR6 and CD161, performed using the data in (B). Representative contour plots comparing Th17 in PBMC and TIL (i). Box and whisker plot comparing the proportion of Th17 among total memory (CD45RA–) non-Treg CD4+ T-cells in PBMC and TIL (ii). G, Quantification of CD4+ Treg cells based on expression of CD25 and CD127, generated using the data in (B). Representative contour plot of Treg cells (CD25+CD127low) from PDAC TIL (i). Box and whisker plot comparing the proportion of Th17 cells in PBMC and TIL (ii). H, Histograms comparing expression levels of activation and differentiation markers on total Tregs from PBMC and TIL, generated using the data in (B). Horizontal lines represent median, boxes represent quartiles and whiskers represent min and max values. Data analyzed using Wilcoxon matched-pairs signed rank test. CyTOF comparisons analyzed using Wilcoxon matched-pairs signed rank test. *, P < 0.05; **, P < 0.01.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/be29/10068448/ee6819e886d6/435fig2.gif)
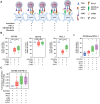
![Figure 3. Characterization of CD8+ T-cell populations within the PDAC tumor microenvironment. A, Dotplot of the top markers expressed in each CD8+ T-cell cluster identified via Louvain clustering of scRNA-seq data (first presented in Fig. 1) from CD8+ T cells (i). UMAP embedding of CD8+ T cells from the 3 PDAC patient samples overlaid with Louvain cluster label (ii). B, CyTOF analysis of CD45+ cells from PDAC patient PBMC and TIL (n = 10), using data first used in Fig. 2B. t-SNE plots show Phenograph-clustered CD8+ T-cell populations in PBMC and TIL. C, Representative contour plot showing CD8+ TRM cells in PDAC TIL based on positive expression of CD69 and CD103, generated using the data in Fig. 2B (i). Box and whisker plot showing the proportion of CD8+ TRM cells in PBMC and TIL, generated using the data in Fig. 2B (ii). D, UMAP embedding, performed using scRNA-seq first presented in Fig. 1, overlaid with module score quintiles and module score distributions by CD8 T-cell cluster from scoring a core module of genes overexpressed in TRM T-cells (i). Differential expression analysis distinguishing TRM-like cells (clusters CD8_1 and CD8_6) from non-TRM cells (ii). Selected genes are labelled, and colored points indicate genes that are differentially expressed [BH adjusted P < 0.01 and absolute (average logFC) > 0.5]. E, Comparison of memory T-cell markers in CD8+ TRM and non-TRM in PDAC TIL, performed using the data in Fig. 2B. Bar graph comparing the proportion of Naïve, EM, CM, and TEMRA subsets in CD8+ TRM and non-TRM cells (i). Bar graph comparing the CD27 and CD28 expression pattern in CD8+ EM T-cells within TRM and non-TRM cells (ii). F, Line graphs comparing T-cell activation and differentiation marker expression on CD8+ TRM versus non-TRM cells in PDAC TIL, generated using the data in Fig. 2B. G, Representative contour plots (i) and quantification (ii) of CD39+ CD8+ TRM cells in PBMC and TIL, generated using the data in Fig. 2B. H, DFS analysis of patients with PDAC from the TCGA-PAAD dataset based on the expression level of ITGAE (CD103) in tumor tissue. Horizontal lines represent median, boxes represent quartiles and whiskers represent min and max values. Data analyzed using Wilcoxon matched-pairs signed rank test. *, P < 0.05; **, P < 0.01.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/be29/10068448/c24913e20497/435fig3.gif)

Similar articles
-
PD-1 and TIGIT coexpressing CD8 + CD103 + tissue-resident memory cells in endometrial cancer as potential targets for immunotherapy.Int Immunopharmacol. 2024 Jan 25;127:111381. doi: 10.1016/j.intimp.2023.111381. Epub 2023 Dec 26. Int Immunopharmacol. 2024. PMID: 38150880
-
The spatial coexistence of TIGIT/CD155 defines poorer survival and resistance to adjuvant chemotherapy in pancreatic ductal adenocarcinoma.Theranostics. 2023 Aug 18;13(13):4601-4614. doi: 10.7150/thno.86547. eCollection 2023. Theranostics. 2023. PMID: 37649613 Free PMC article.
-
TIGIT Expression Delineates T-cell Populations with Distinct Functional and Prognostic Impact in Pancreatic Cancer.Clin Cancer Res. 2023 Jul 14;29(14):2638-2650. doi: 10.1158/1078-0432.CCR-23-0258. Clin Cancer Res. 2023. PMID: 37140899 Free PMC article.
-
TIGIT in cancer immunotherapy.J Immunother Cancer. 2020 Sep;8(2):e000957. doi: 10.1136/jitc-2020-000957. J Immunother Cancer. 2020. PMID: 32900861 Free PMC article. Review.
-
The Emerging Role of CD8+ Tissue Resident Memory T (TRM) Cells in Antitumor Immunity: A Unique Functional Contribution of the CD103 Integrin.Front Immunol. 2018 Aug 15;9:1904. doi: 10.3389/fimmu.2018.01904. eCollection 2018. Front Immunol. 2018. PMID: 30158938 Free PMC article. Review.
Cited by
-
Tissue-Resident Memory T Cells in Gastrointestinal Cancers: Prognostic Significance and Therapeutic Implications.Biomedicines. 2024 Jun 17;12(6):1342. doi: 10.3390/biomedicines12061342. Biomedicines. 2024. PMID: 38927549 Free PMC article. Review.
-
Plasticity and Tumor Microenvironment in Pancreatic Cancer: Genetic, Metabolic, and Immune Perspectives.Cancers (Basel). 2024 Dec 6;16(23):4094. doi: 10.3390/cancers16234094. Cancers (Basel). 2024. PMID: 39682280 Free PMC article. Review.
-
TIGIT blockade in the context of BCMA-CART cell therapy does not augment efficacy in a multiple myeloma mouse model.Oncoimmunology. 2025 Dec;14(1):2529632. doi: 10.1080/2162402X.2025.2529632. Epub 2025 Jul 12. Oncoimmunology. 2025. PMID: 40652308 Free PMC article.
-
Expression and role of the immune checkpoint regulator PD-L1 in the tumor-stroma interplay of pancreatic ductal adenocarcinoma.Front Immunol. 2023 Jun 28;14:1157397. doi: 10.3389/fimmu.2023.1157397. eCollection 2023. Front Immunol. 2023. PMID: 37449210 Free PMC article.
-
Single cell RNA-sequencing delineates CD8+ tissue resident memory T cells maintaining rejection in liver transplantation.Theranostics. 2024 Aug 12;14(12):4844-4860. doi: 10.7150/thno.96928. eCollection 2024. Theranostics. 2024. PMID: 39239518 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

