The transcription factor ATF3 switches cell death from apoptosis to necroptosis in hepatic steatosis in male mice
- PMID: 36690638
- PMCID: PMC9871012
- DOI: 10.1038/s41467-023-35804-w
The transcription factor ATF3 switches cell death from apoptosis to necroptosis in hepatic steatosis in male mice
Abstract
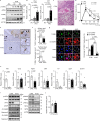
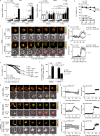
Hepatocellular death increases with hepatic steatosis aggravation, although its regulation remains unclear. Here we show that hepatic steatosis aggravation shifts the hepatocellular death mode from apoptosis to necroptosis, causing increased hepatocellular death. Our results reveal that the transcription factor ATF3 acts as a master regulator in this shift by inducing expression of RIPK3, a regulator of necroptosis. In severe hepatic steatosis, after partial hepatectomy, hepatic ATF3-deficient or -overexpressing mice display decreased or increased RIPK3 expression and necroptosis, respectively. In cultured hepatocytes, ATF3 changes TNFα-dependent cell death mode from apoptosis to necroptosis, as revealed by live-cell imaging. In non-alcoholic steatohepatitis (NASH) mice, hepatic ATF3 deficiency suppresses RIPK3 expression and hepatocellular death. In human NASH, hepatocellular damage is correlated with the frequency of hepatocytes expressing ATF3 or RIPK3, which overlap frequently. ATF3-dependent RIPK3 induction, causing a modal shift of hepatocellular death, can be a therapeutic target for steatosis-induced liver damage, including NASH.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








Similar articles
-
Necroptosis is a key pathogenic event in human and experimental murine models of non-alcoholic steatohepatitis.Clin Sci (Lond). 2015 Oct 1;129(8):721-39. doi: 10.1042/CS20140732. Epub 2015 Jun 15. Clin Sci (Lond). 2015. PMID: 26201023
-
Inhibition of receptor-interacting protein kinase 1 improves experimental non-alcoholic fatty liver disease.J Hepatol. 2020 Apr;72(4):627-635. doi: 10.1016/j.jhep.2019.11.008. Epub 2019 Nov 21. J Hepatol. 2020. PMID: 31760070
-
Inhibition of ZFP281/ZNF281-RIPK1/RIPK3/MLKL signaling in hepatocytes by pterostilbene relieves hepatic lipometabolic disorder and inflammation in non-alcoholic steatohepatitis.Int Immunopharmacol. 2025 Jan 27;146:113936. doi: 10.1016/j.intimp.2024.113936. Epub 2024 Dec 25. Int Immunopharmacol. 2025. PMID: 39724734
-
RIPK1 and RIPK3: critical regulators of inflammation and cell death.Trends Cell Biol. 2015 Jun;25(6):347-53. doi: 10.1016/j.tcb.2015.01.001. Epub 2015 Feb 4. Trends Cell Biol. 2015. PMID: 25662614 Review.
-
Mechanisms of TNF-independent RIPK3-mediated cell death.Biochem J. 2022 Oct 14;479(19):2049-2062. doi: 10.1042/BCJ20210724. Biochem J. 2022. PMID: 36240069 Free PMC article. Review.
Cited by
-
Relationship Between Activating Transcription Factor 3 and Forkhead Box Protein A2 in Spinal Cord Injury and the Underlying Mechanism.Appl Biochem Biotechnol. 2025 Aug;197(8):5327-5343. doi: 10.1007/s12010-025-05256-7. Epub 2025 Jun 4. Appl Biochem Biotechnol. 2025. PMID: 40464835
-
Medaka liver developed Human NAFLD-NASH transcriptional signatures in response to ancestral bisphenol A exposure.Res Sq [Preprint]. 2024 Jul 16:rs.3.rs-4585175. doi: 10.21203/rs.3.rs-4585175/v1. Res Sq. 2024. PMID: 39070641 Free PMC article. Preprint.
-
Killing hepatocellular carcinoma in the NAFLD/NASH stage: a comprehensive perspective on targeting regulated cell death.Cell Death Discov. 2025 Jun 19;11(1):281. doi: 10.1038/s41420-025-02558-x. Cell Death Discov. 2025. PMID: 40537476 Free PMC article. Review.
-
Bibliometric analysis of autophagy in NAFLD from 2004 to 2023.Medicine (Baltimore). 2024 Dec 6;103(49):e40835. doi: 10.1097/MD.0000000000040835. Medicine (Baltimore). 2024. PMID: 39654183 Free PMC article.
-
METTL3 restricts RIPK1-dependent cell death via the ATF3-cFLIP axis in the intestinal epithelium.Cell Regen. 2024 Aug 2;13(1):14. doi: 10.1186/s13619-024-00197-8. Cell Regen. 2024. PMID: 39093347 Free PMC article.
References
-
- Campana, L., Esser, H., Huch, M. & Forbes, S. Liver regeneration and inflammation: from fundamental science to clinical applications. Nat. Rev. Mol. Cell Biol.22, 608–524 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

