Predicting severity in COVID-19 disease using sepsis blood gene expression signatures
- PMID: 36690713
- PMCID: PMC9868505
- DOI: 10.1038/s41598-023-28259-y
Predicting severity in COVID-19 disease using sepsis blood gene expression signatures
Abstract
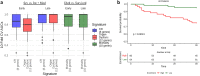
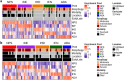
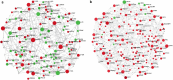
Severely-afflicted COVID-19 patients can exhibit disease manifestations representative of sepsis, including acute respiratory distress syndrome and multiple organ failure. We hypothesized that diagnostic tools used in managing all-cause sepsis, such as clinical criteria, biomarkers, and gene expression signatures, should extend to COVID-19 patients. Here we analyzed the whole blood transcriptome of 124 early (1-5 days post-hospital admission) and late (6-20 days post-admission) sampled patients with confirmed COVID-19 infections from hospitals in Quebec, Canada. Mechanisms associated with COVID-19 severity were identified between severity groups (ranging from mild disease to the requirement for mechanical ventilation and mortality), and established sepsis signatures were assessed for dysregulation. Specifically, gene expression signatures representing pathophysiological events, namely cellular reprogramming, organ dysfunction, and mortality, were significantly enriched and predictive of severity and lethality in COVID-19 patients. Mechanistic endotypes reflective of distinct sepsis aetiologies and therapeutic opportunities were also identified in subsets of patients, enabling prediction of potentially-effective repurposed drugs. The expression of sepsis gene expression signatures in severely-afflicted COVID-19 patients indicates that these patients should be classified as having severe sepsis. Accordingly, in severe COVID-19 patients, these signatures should be strongly considered for the mechanistic characterization, diagnosis, and guidance of treatment using repurposed drugs.
© 2023. The Author(s).
Conflict of interest statement
RH has filed the CR signature for patent protection and licenced this to Asep Medical, a Vancouver company in which he has a significant ownership position. RH, GCF, and AB have filed the all-cause sepsis endotype signatures for patent protection. Other authors have no competing interests to declare.
Figures

Similar articles
-
Predicting sepsis severity at first clinical presentation: The role of endotypes and mechanistic signatures.EBioMedicine. 2022 Jan;75:103776. doi: 10.1016/j.ebiom.2021.103776. Epub 2022 Jan 10. EBioMedicine. 2022. PMID: 35027333 Free PMC article.
-
Prospective validation of a transcriptomic severity classifier among patients with suspected acute infection and sepsis in the emergency department.Eur J Emerg Med. 2022 Oct 1;29(5):357-365. doi: 10.1097/MEJ.0000000000000931. Epub 2022 Apr 21. Eur J Emerg Med. 2022. PMID: 35467566 Free PMC article.
-
Targeted plasma proteomics reveals signatures discriminating COVID-19 from sepsis with pneumonia.Respir Res. 2023 Feb 24;24(1):62. doi: 10.1186/s12931-023-02364-y. Respir Res. 2023. PMID: 36829233 Free PMC article.
-
Insights into COVID-19-associated critical illness: a narrative review.Ann Transl Med. 2023 Mar 15;11(5):220. doi: 10.21037/atm-22-2541. Epub 2023 Feb 16. Ann Transl Med. 2023. PMID: 37007577 Free PMC article. Review.
-
A Comparison between SARS-CoV-2 and Gram-Negative Bacteria-Induced Hyperinflammation and Sepsis.Int J Mol Sci. 2023 Oct 14;24(20):15169. doi: 10.3390/ijms242015169. Int J Mol Sci. 2023. PMID: 37894850 Free PMC article. Review.
Cited by
-
Facilitating systems-level analyses of all-cause and Covid-mediated sepsis through SeptiSearch, a manually-curated compendium of dysregulated gene sets.Front Immunol. 2023 May 26;14:1135859. doi: 10.3389/fimmu.2023.1135859. eCollection 2023. Front Immunol. 2023. PMID: 37304268 Free PMC article.
-
Minimalistic Transcriptomic Signatures Permit Accurate Early Prediction of COVID-19 Mortality.medRxiv [Preprint]. 2025 May 19:2025.05.18.25327658. doi: 10.1101/2025.05.18.25327658. medRxiv. 2025. PMID: 40475132 Free PMC article. Preprint.
-
The Impact of COVID-19 on Sepsis-Related Mortality in the United States.J Clin Med Res. 2023 Jun;15(6):328-331. doi: 10.14740/jocmr4937. Epub 2023 Jun 29. J Clin Med Res. 2023. PMID: 37434769 Free PMC article.
-
Biomarkers to guide sepsis management.Ann Intensive Care. 2025 Jul 21;15(1):103. doi: 10.1186/s13613-025-01524-1. Ann Intensive Care. 2025. PMID: 40685448 Free PMC article. Review.
-
COVID-19-Associated Sepsis: Potential Role of Phytochemicals as Functional Foods and Nutraceuticals.Int J Mol Sci. 2024 Aug 3;25(15):8481. doi: 10.3390/ijms25158481. Int J Mol Sci. 2024. PMID: 39126050 Free PMC article. Review.
References
-
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Yu T. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

