Precise transcript targeting by CRISPR-Csm complexes
- PMID: 36690762
- PMCID: PMC10497410
- DOI: 10.1038/s41587-022-01649-9
Precise transcript targeting by CRISPR-Csm complexes
Abstract
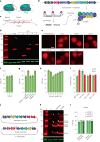
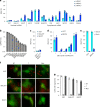
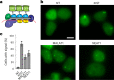
Robust and precise transcript targeting in mammalian cells remains a difficult challenge using existing approaches due to inefficiency, imprecision and subcellular compartmentalization. Here we show that the clustered regularly interspaced short palindromic repeats (CRISPR)-Csm complex, a multiprotein effector from type III CRISPR immune systems in prokaryotes, provides surgical RNA ablation of both nuclear and cytoplasmic transcripts. As part of the most widely occurring CRISPR adaptive immune pathway, CRISPR-Csm uses a programmable RNA-guided mechanism to find and degrade target RNA molecules without inducing indiscriminate trans-cleavage of cellular RNAs, giving it an important advantage over the CRISPR-Cas13 family of enzymes. Using single-vector delivery of the Streptococcus thermophilus Csm complex, we observe high-efficiency RNA knockdown (90-99%) and minimal off-target effects in human cells, outperforming existing technologies including short hairpin RNA- and Cas13-mediated knockdown. We also find that catalytically inactivated Csm achieves specific and durable RNA binding, a property we harness for live-cell RNA imaging. These results establish the feasibility and efficacy of multiprotein CRISPR-Cas effector complexes as RNA-targeting tools in eukaryotes.
© 2023. The Author(s).
Conflict of interest statement
The Regents of the University of California have filed a related patent, for which D.C. and J.A.D. are inventors, on the use of the Csm system for eukaryotic RNA KD with the United States Patent and Trademark Office. J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics and Mammoth Biosciences, and a director of Johnson & Johnson, Altos Labs and Tempus. J.A.D. is a scientific advisor to Caribou Biosciences, Intellia Therapeutics, Mammoth Biosciences, Scribe Therapeutics, Algen, Felix and Inari; J.A.D. serves on the scientific advisory boards of Sixth Street and The Column Group. J.A.D. has research projects sponsored by Roche and Apple Tree Partners. The other author declares no competing interests.
Figures

Comment in
-
CRISPR-Csm for eukaryotic RNA knockdown and imaging without toxicity.Nat Biotechnol. 2023 Sep;41(9):1204-1205. doi: 10.1038/s41587-023-01665-3. Nat Biotechnol. 2023. PMID: 36693989 No abstract available.
Similar articles
-
Optimization of specific RNA knockdown in mammalian cells with CRISPR-Cas13.Methods. 2022 Oct;206:58-68. doi: 10.1016/j.ymeth.2022.08.007. Epub 2022 Aug 17. Methods. 2022. PMID: 35987443 Free PMC article.
-
Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.Mol Cell. 2019 Jan 17;73(2):264-277.e5. doi: 10.1016/j.molcel.2018.11.007. Epub 2018 Nov 29. Mol Cell. 2019. PMID: 30503773 Free PMC article.
-
RNA-Targeting CRISPR-Cas Systems and Their Applications.Int J Mol Sci. 2020 Feb 7;21(3):1122. doi: 10.3390/ijms21031122. Int J Mol Sci. 2020. PMID: 32046217 Free PMC article. Review.
-
Target preference of Type III-A CRISPR-Cas complexes at the transcription bubble.Nat Commun. 2019 Jul 5;10(1):3001. doi: 10.1038/s41467-019-10780-2. Nat Commun. 2019. PMID: 31278272 Free PMC article.
-
Progress of CRISPR-based programmable RNA manipulation and detection.Wiley Interdiscip Rev RNA. 2023 Nov-Dec;14(6):e1804. doi: 10.1002/wrna.1804. Epub 2023 Jun 6. Wiley Interdiscip Rev RNA. 2023. PMID: 37282821 Review.
Cited by
-
Approaches to probe and perturb long noncoding RNA functions in diseases.Curr Opin Genet Dev. 2024 Apr;85:102158. doi: 10.1016/j.gde.2024.102158. Epub 2024 Feb 26. Curr Opin Genet Dev. 2024. PMID: 38412563 Free PMC article. Review.
-
Probing the orthogonality and robustness of the mammalian RNA-binding protein Musashi-1 in Escherichia coli.J Biol Eng. 2024 Sep 30;18(1):52. doi: 10.1186/s13036-024-00448-x. J Biol Eng. 2024. PMID: 39350178 Free PMC article.
-
Beyond ribosome biogenesis: noncoding nucleolar RNAs in physiology and tumor biology.Nucleus. 2023 Dec;14(1):2274655. doi: 10.1080/19491034.2023.2274655. Epub 2023 Oct 31. Nucleus. 2023. PMID: 37906621 Free PMC article. Review.
-
Engineered smart materials for RNA based molecular therapy to treat Glioblastoma.Bioact Mater. 2023 Nov 27;33:396-423. doi: 10.1016/j.bioactmat.2023.11.007. eCollection 2024 Mar. Bioact Mater. 2023. PMID: 38059120 Free PMC article.
-
A tightly controlled gene induction system that contributes to the study of lethal gene function in Streptococcus mutans.J Oral Microbiol. 2023 Sep 7;15(1):2253675. doi: 10.1080/20002297.2023.2253675. eCollection 2023. J Oral Microbiol. 2023. PMID: 37691880 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

