TRIM24 controls induction of latent HIV-1 by stimulating transcriptional elongation
- PMID: 36690785
- PMCID: PMC9870992
- DOI: 10.1038/s42003-023-04484-z
TRIM24 controls induction of latent HIV-1 by stimulating transcriptional elongation
Abstract
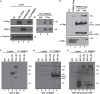
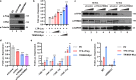
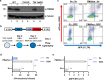
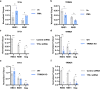
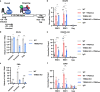
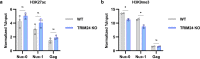
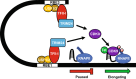
Binding of USF1/2 and TFII-I (RBF-2) at conserved sites flanking the HIV-1 LTR enhancer is essential for reactivation from latency in T cells, with TFII-I knockdown rendering the provirus insensitive to T cell signaling. We identified an interaction of TFII-I with the tripartite motif protein TRIM24, and these factors were found to be constitutively associated with the HIV-1 LTR. Similar to the effect of TFII-I depletion, loss of TRIM24 impaired reactivation of HIV-1 in response to T cell signaling. TRIM24 deficiency did not affect recruitment of RNA Pol II to the LTR promoter, but inhibited transcriptional elongation, an effect that was associated with decreased RNA Pol II CTD S2 phosphorylation and impaired recruitment of CDK9. A considerable number of genomic loci are co-occupied by TRIM24/TFII-I, and we found that TRIM24 deletion caused altered T cell immune response, an effect that is facilitated by TFII-I. These results demonstrate a role of TRIM24 for regulation of transcriptional elongation from the HIV-1 promoter, through its interaction with TFII-I, and by recruitment of P-TEFb. Furthermore, these factors co-regulate a significant proportion of genes involved in T cell immune response, consistent with tight coupling of HIV-1 transcriptional activation and T cell signaling.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Inhibition of the TRIM24 bromodomain reactivates latent HIV-1.Sci Rep. 2023 Jan 11;13(1):556. doi: 10.1038/s41598-023-27765-3. Sci Rep. 2023. PMID: 36631514 Free PMC article.
-
Upstream Stimulatory Factors Regulate HIV-1 Latency and Are Required for Robust T Cell Activation.Viruses. 2023 Jun 28;15(7):1470. doi: 10.3390/v15071470. Viruses. 2023. PMID: 37515158 Free PMC article.
-
Short Communication: The Broad-Spectrum Histone Deacetylase Inhibitors Vorinostat and Panobinostat Activate Latent HIV in CD4(+) T Cells In Part Through Phosphorylation of the T-Loop of the CDK9 Subunit of P-TEFb.AIDS Res Hum Retroviruses. 2016 Feb;32(2):169-73. doi: 10.1089/AID.2015.0347. AIDS Res Hum Retroviruses. 2016. PMID: 26727990 Free PMC article.
-
Efficient Non-Epigenetic Activation of HIV Latency through the T-Cell Receptor Signalosome.Viruses. 2020 Aug 8;12(8):868. doi: 10.3390/v12080868. Viruses. 2020. PMID: 32784426 Free PMC article. Review.
-
The HIV-1 Tat Protein: Mechanism of Action and Target for HIV-1 Cure Strategies.Curr Pharm Des. 2017;23(28):4098-4102. doi: 10.2174/1381612823666170704130635. Curr Pharm Des. 2017. PMID: 28677507 Free PMC article. Review.
Cited by
-
Release of P-TEFb from the Super Elongation Complex promotes HIV-1 latency reversal.bioRxiv [Preprint]. 2024 Mar 1:2024.03.01.582881. doi: 10.1101/2024.03.01.582881. bioRxiv. 2024. Update in: PLoS Pathog. 2024 Sep 11;20(9):e1012083. doi: 10.1371/journal.ppat.1012083. PMID: 38464055 Free PMC article. Updated. Preprint.
-
Release of P-TEFb from the Super Elongation Complex promotes HIV-1 latency reversal.PLoS Pathog. 2024 Sep 11;20(9):e1012083. doi: 10.1371/journal.ppat.1012083. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39259751 Free PMC article.
-
Comprehensive SUMO Proteomic Analyses Identify HIV Latency-Associated Proteins in Microglia.Cells. 2025 Feb 6;14(3):235. doi: 10.3390/cells14030235. Cells. 2025. PMID: 39937027 Free PMC article.
-
CDK8 inhibitors antagonize HIV-1 reactivation and promote provirus latency in T cells.J Virol. 2023 Sep 28;97(9):e0092323. doi: 10.1128/jvi.00923-23. Epub 2023 Sep 6. J Virol. 2023. PMID: 37671866 Free PMC article.
-
The cell biology of HIV-1 latency and rebound.Retrovirology. 2024 Apr 5;21(1):6. doi: 10.1186/s12977-024-00639-w. Retrovirology. 2024. PMID: 38580979 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

