Dinuclear complex-induced DNA melting
- PMID: 36691056
- PMCID: PMC9869567
- DOI: 10.1186/s12951-023-01784-8
Dinuclear complex-induced DNA melting
Abstract
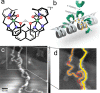
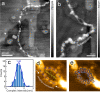
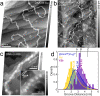
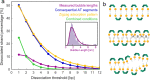
Dinuclear copper complexes have been designed for molecular recognition in order to selectively bind to two neighboring phosphate moieties in the backbone of double strand DNA. Associated biophysical, biochemical and cytotoxic effects on DNA were investigated in previous works, where atomic force microscopy (AFM) in ambient conditions turned out to be a particular valuable asset, since the complexes influence the macromechanical properties and configurations of the strands. To investigate and scrutinize these effects in more depth from a structural point of view, cutting-edge preparation methods and scanning force microscopy under ultra-high vacuum (UHV) conditions were employed to yield submolecular resolution images. DNA strand mechanics and interactions could be resolved on the single base pair level, including the amplified formation of melting bubbles. Even the interaction of singular complex molecules could be observed. To better assess the results, the appearance of treated DNA is also compared to the behavior of untreated DNA in UHV on different substrates. Finally, we present data from a statistical simulation reasoning about the nanomechanics of strand dissociation. This sort of quantitative experimental insights paralleled by statistical simulations impressively shade light on the rationale for strand dissociations of this novel DNA interaction process, that is an important nanomechanistic key and novel approach for the development of new chemotherapeutic agents.
Keywords: AFM; Biomolecules; DNA; Electrospray ionization; Molecular recognition.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy.Acc Chem Res. 2014 Jun 17;47(6):1645-53. doi: 10.1021/ar400299m. Epub 2014 Mar 6. Acc Chem Res. 2014. PMID: 24601497
-
Different pulling modes in DNA overstretching: a theoretical analysis.Phys Rev E Stat Nonlin Soft Matter Phys. 2010 May;81(5 Pt 1):051926. doi: 10.1103/PhysRevE.81.051926. Epub 2010 May 27. Phys Rev E Stat Nonlin Soft Matter Phys. 2010. PMID: 20866280
-
Raman spectroscopy of DNA-metal complexes. II. The thermal denaturation of DNA in the presence of Sr2+, Ba2+, Mg2+, Ca2+, Mn2+, Co2+, Ni2+, and Cd2+.Biophys J. 1995 Dec;69(6):2623-41. doi: 10.1016/S0006-3495(95)80133-5. Biophys J. 1995. PMID: 8599669 Free PMC article.
-
DNA-directed base pair opening.Molecules. 2012 Oct 11;17(10):11947-64. doi: 10.3390/molecules171011947. Molecules. 2012. PMID: 23060287 Free PMC article. Review.
-
DNA substrate preparation for atomic force microscopy studies of protein-DNA interactions.J Mol Recognit. 2013 Dec;26(12):605-17. doi: 10.1002/jmr.2311. J Mol Recognit. 2013. PMID: 24277605 Review.
References
-
- Jany T, Moreth A, Gruschka C, Sischka A, Spiering A, Dieding M, Wang Y, Samo SH, Stammler A, Bögge H, von Fischer Mollard G, Anselmetti D, Glaser T. Rational design of a cytotoxic dinuclear Cu2 complex that binds by molecular recognition at two neighboring phosphates of the DNA backbone. Inorg Chem. 2015;54:2679–2690. doi: 10.1021/ic5028465. - DOI - PubMed
-
- Jany T, Horstmann née Gruschka C, Bögge H, Stammler A, Glaser T. A series of dinuclear complexes with a flexible naphthalene-spacer and MOM-cleavage by pre-coordinated lewis acids. Z Anorg Allg Chem. 2015;641:2157–2168. doi: 10.1002/zaac.201500553. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

