An atlas of lamina-associated chromatin across twelve human cell types reveals an intermediate chromatin subtype
- PMID: 36691074
- PMCID: PMC9869549
- DOI: 10.1186/s13059-023-02849-5
An atlas of lamina-associated chromatin across twelve human cell types reveals an intermediate chromatin subtype
Abstract
Background: Association of chromatin with lamin proteins at the nuclear periphery has emerged as a potential mechanism to coordinate cell type-specific gene expression and maintain cellular identity via gene silencing. Unlike many histone modifications and chromatin-associated proteins, lamina-associated domains (LADs) are mapped genome-wide in relatively few genetically normal human cell types, which limits our understanding of the role peripheral chromatin plays in development and disease.
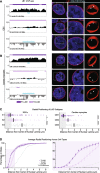
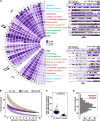
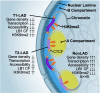
Results: To address this gap, we map LAMIN B1 occupancy across twelve human cell types encompassing pluripotent stem cells, intermediate progenitors, and differentiated cells from all three germ layers. Integrative analyses of this atlas with gene expression and repressive histone modification maps reveal that lamina-associated chromatin in all twelve cell types is organized into at least two subtypes defined by differences in LAMIN B1 occupancy, gene expression, chromatin accessibility, transposable elements, replication timing, and radial positioning. Imaging of fluorescently labeled DNA in single cells validates these subtypes and shows radial positioning of LADs with higher LAMIN B1 occupancy and heterochromatic histone modifications primarily embedded within the lamina. In contrast, the second subtype of lamina-associated chromatin is relatively gene dense, accessible, dynamic across development, and positioned adjacent to the lamina. Most genes gain or lose LAMIN B1 occupancy consistent with cell types along developmental trajectories; however, we also identify examples where the enhancer, but not the gene body and promoter, changes LAD state.
Conclusions: Altogether, this atlas represents the largest resource to date for peripheral chromatin organization studies and reveals an intermediate chromatin subtype.
Keywords: 3D genome; Cellular differentiation; Lamina-associated domains; Peripheral chromatin organization.
© 2023. The Author(s).
Conflict of interest statement
K.C.K. is currently an employee of FaunaBio. A.C. is currently an employee of Orca Bio. K.S.P. is a consultant for Tenaya Therapeutics.
Figures




Similar articles
-
Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery.Mol Cell. 2018 Sep 6;71(5):802-815.e7. doi: 10.1016/j.molcel.2018.05.017. Epub 2018 Jun 28. Mol Cell. 2018. PMID: 30201095 Free PMC article.
-
Gene Regulatory Interactions at Lamina-Associated Domains.Genes (Basel). 2023 Jan 28;14(2):334. doi: 10.3390/genes14020334. Genes (Basel). 2023. PMID: 36833261 Free PMC article.
-
Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin.Nucleus. 2015;6(1):30-9. doi: 10.4161/19491034.2014.990855. Nucleus. 2015. PMID: 25602132 Free PMC article.
-
Lamina-associated domains: peripheral matters and internal affairs.Genome Biol. 2020 Apr 2;21(1):85. doi: 10.1186/s13059-020-02003-5. Genome Biol. 2020. PMID: 32241294 Free PMC article. Review.
-
The interplay between histone modifications and nuclear lamina in genome regulation.J Genet Genomics. 2025 Jan;52(1):24-38. doi: 10.1016/j.jgg.2024.10.005. Epub 2024 Oct 18. J Genet Genomics. 2025. PMID: 39426590 Review.
Cited by
-
CRWN nuclear lamina components maintain the H3K27me3 landscape and promote successful reproduction in Arabidopsis.bioRxiv [Preprint]. 2023 Oct 3:2023.10.03.560721. doi: 10.1101/2023.10.03.560721. bioRxiv. 2023. Update in: New Phytol. 2024 Jul;243(1):213-228. doi: 10.1111/nph.19791. PMID: 37873406 Free PMC article. Updated. Preprint.
-
Major nuclear locales define nuclear genome organization and function beyond A and B compartments.Elife. 2025 Apr 25;13:RP99116. doi: 10.7554/eLife.99116. Elife. 2025. PMID: 40279158 Free PMC article.
-
LAP2alpha facilitates myogenic gene expression by preventing nucleoplasmic lamin A/C from spreading to active chromatin regions.Nucleic Acids Res. 2024 Oct 28;52(19):11500-11518. doi: 10.1093/nar/gkae752. Nucleic Acids Res. 2024. PMID: 39228367 Free PMC article.
-
Depletion of lamins B1 and B2 promotes chromatin mobility and induces differential gene expression by a mesoscale-motion-dependent mechanism.Genome Biol. 2024 Mar 22;25(1):77. doi: 10.1186/s13059-024-03212-y. Genome Biol. 2024. PMID: 38519987 Free PMC article.
-
A screen for regeneration-associated silencer regulatory elements in zebrafish.Dev Cell. 2024 Mar 11;59(5):676-691.e5. doi: 10.1016/j.devcel.2024.01.004. Epub 2024 Jan 29. Dev Cell. 2024. PMID: 38290519 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL098179/HL/NHLBI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U01 HL157989/HL/NHLBI NIH HHS/United States
- F31 HL160114/HL/NHLBI NIH HHS/United States
- UM1 HL098179/HL/NHLBI NIH HHS/United States
- F31 HL160114/NH/NIH HHS/United States
- R01 HL134956/HL/NHLBI NIH HHS/United States
- HL147123/NH/NIH HHS/United States
- U01 DA052715/DA/NIDA NIH HHS/United States
- DP2 HL147123/HL/NHLBI NIH HHS/United States
- GM140324/NH/NIH HHS/United States
- AHA/American Heart Association-American Stroke Association/United States
- R01 GM137425/GM/NIGMS NIH HHS/United States
- TL1TR001880/TR/NCATS NIH HHS/United States
- TL1 TR001880/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

