Digital twin demonstrates significance of biomechanical growth control in liver regeneration after partial hepatectomy
- PMID: 36691615
- PMCID: PMC9860368
- DOI: 10.1016/j.isci.2022.105714
Digital twin demonstrates significance of biomechanical growth control in liver regeneration after partial hepatectomy
Abstract
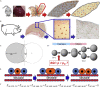
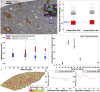
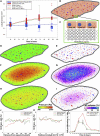
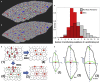
Partial liver removal is an important therapy option for liver cancer. In most patients within a few weeks, the liver is able to fully regenerate. In some patients, however, regeneration fails with often severe consequences. To better understand the control mechanisms of liver regeneration, experiments in mice were performed, guiding the creation of a spatiotemporal 3D model of the regenerating liver. The model represents cells and blood vessels within an entire liver lobe, a macroscopic liver subunit. The model could reproduce the experimental data only if a biomechanical growth control (BGC)-mechanism, inhibiting cell cycle entrance at high compression, was taken into account and predicted that BGC may act as a short-range growth inhibitor minimizing the number of proliferating neighbor cells of a proliferating cell, generating a checkerboard-like proliferation pattern. Model-predicted cell proliferation patterns in pigs and mice were found experimentally. The results underpin the importance of biomechanical aspects in liver growth control.
Keywords: Tissue engineering; mathematical biosciences; systems biology.
© 2022.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
A liver digital twin for in silico testing of cellular and inter-cellular mechanisms in regeneration after drug-induced damage.iScience. 2023 Sep 28;27(2):108077. doi: 10.1016/j.isci.2023.108077. eCollection 2024 Feb 16. iScience. 2023. PMID: 38371522 Free PMC article.
-
Endothelial-directed hepatic regeneration after partial hepatectomy.Ann Surg. 2003 Apr;237(4):530-5. doi: 10.1097/01.SLA.0000059986.96051.EA. Ann Surg. 2003. PMID: 12677150 Free PMC article.
-
The impact of the intra-abdominal space on liver regeneration after a partial hepatectomy in rats.J Surg Res. 2011 Nov;171(1):259-65. doi: 10.1016/j.jss.2010.01.012. Epub 2010 Feb 4. J Surg Res. 2011. PMID: 20421115
-
The vascular endothelial growth factor signaling pathway regulates liver sinusoidal endothelial cells during liver regeneration after partial hepatectomy.Expert Rev Gastroenterol Hepatol. 2021 Feb;15(2):139-147. doi: 10.1080/17474124.2020.1815532. Epub 2020 Oct 14. Expert Rev Gastroenterol Hepatol. 2021. PMID: 32902336 Review.
-
Metallothionein and liver cell regeneration.Exp Biol Med (Maywood). 2006 Feb;231(2):138-44. doi: 10.1177/153537020623100203. Exp Biol Med (Maywood). 2006. PMID: 16446489 Review.
Cited by
-
A liver digital twin for in silico testing of cellular and inter-cellular mechanisms in regeneration after drug-induced damage.iScience. 2023 Sep 28;27(2):108077. doi: 10.1016/j.isci.2023.108077. eCollection 2024 Feb 16. iScience. 2023. PMID: 38371522 Free PMC article.
-
The evolution and revolution of artificial intelligence in hepatology: From current applications to future paradigms.Hepatol Forum. 2024 Jul 2;5(3):97-99. doi: 10.14744/hf.2024.2024.ed0001. eCollection 2024. Hepatol Forum. 2024. PMID: 39006141 Free PMC article. No abstract available.
-
Cross-species variability in lobular geometry and cytochrome P450 hepatic zonation: insights into CYP1A2, CYP2D6, CYP2E1 and CYP3A4.Front Pharmacol. 2024 May 16;15:1404938. doi: 10.3389/fphar.2024.1404938. eCollection 2024. Front Pharmacol. 2024. PMID: 38818378 Free PMC article.
-
From sampling to simulating: Single-cell multiomics in systems pathophysiological modeling.iScience. 2024 Nov 5;27(12):111322. doi: 10.1016/j.isci.2024.111322. eCollection 2024 Dec 20. iScience. 2024. PMID: 39628578 Free PMC article. Review.
-
Phenotypic characterization of liver tissue heterogeneity through a next-generation 3D single-cell atlas.Sci Rep. 2024 Feb 3;14(1):2823. doi: 10.1038/s41598-024-53309-4. Sci Rep. 2024. PMID: 38307948 Free PMC article.
References
-
- Alessandri K., Sarangi B.R., Gurchenkov V.V., Sinha B., Kießling T.R., Fetler L., et al. Cellular capsules as a tool for multicellular spheroid production and for investigating the mechanics of tumor progression in vitro. Proc. Natl. Acad. Sci. USA. 2013;110:14843–14848. doi: 10.1073/pnas.1309482110. - DOI - PMC - PubMed
-
- Almet A.A., Maini P.K., Moulton D.E., Byrne H.M. Modeling perspectives on the intestinal crypt, a canonical system for growth, mechanics, and remodeling. Curr. Opin. Biomed. Eng. 2020;15:32–39.
-
- Ambrosi D., Preziosi L., Vitale G. The interplay between stress and growth in solid tumors. Mech. Res. Commun. 2012;42:87–91. doi: 10.1016/J.MECHRESCOM.2012.01.002. - DOI
-
- Anderson A.R.A., Chaplain M.A.J., Rejniak K.A., editors. Single-cell-based models in biology and medicine. Birkhäuser; 2007.
LinkOut - more resources
Full Text Sources

