Attenuating iPSC reprogramming stress with dominant-negative BET peptides
- PMID: 36691621
- PMCID: PMC9860338
- DOI: 10.1016/j.isci.2022.105889
Attenuating iPSC reprogramming stress with dominant-negative BET peptides
Abstract
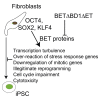
Generation of induced pluripotent stem cells (iPSCs) is inefficient and stochastic. The underlying causes for these deficiencies are elusive. Here, we showed that the reprogramming factors (OCT4, SOX2, and KLF4, collectively OSK) elicit dramatic reprogramming stress even without the pro-oncogene MYC including massive transcriptional turbulence, massive and random deregulation of stress-response genes, cell cycle impairment, downregulation of mitotic genes, illegitimate reprogramming, and cytotoxicity. The conserved dominant-negative (DN) peptides of the three ubiquitous human bromodomain and extraterminal (BET) proteins enhanced iPSC reprogramming and mitigated all the reprogramming stresses mentioned above. The concept of reprogramming stress developed here affords an alternative avenue to understanding and improving iPSC reprogramming. These DN BET fragments target a similar set of the genes as the BET chemical inhibitors do, indicating a distinct approach to targeting BET proteins.
Keywords: Peptides; Protein; Stem cell plasticity; Stem cells research.
© 2022 The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures






Similar articles
-
Enhanced human somatic cell reprogramming efficiency by fusion of the MYC transactivation domain and OCT4.Stem Cell Res. 2017 Dec;25:88-97. doi: 10.1016/j.scr.2017.10.014. Epub 2017 Oct 26. Stem Cell Res. 2017. PMID: 29125994
-
Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway.Cell Cycle. 2013 Sep 15;12(18):3109-24. doi: 10.4161/cc.26173. Epub 2013 Aug 21. Cell Cycle. 2013. PMID: 23974095 Free PMC article.
-
Quick, Coordinated and Authentic Reprogramming of Ribosome Biogenesis during iPSC Reprogramming.Cells. 2020 Nov 15;9(11):2484. doi: 10.3390/cells9112484. Cells. 2020. PMID: 33203179 Free PMC article.
-
Inducing pluripotency in vitro: recent advances and highlights in induced pluripotent stem cells generation and pluripotency reprogramming.Cell Prolif. 2015 Apr;48(2):140-56. doi: 10.1111/cpr.12162. Epub 2015 Jan 29. Cell Prolif. 2015. PMID: 25643745 Free PMC article. Review.
-
Induced pluripotent stem cell technology: trends in molecular biology, from genetics to epigenetics.Epigenomics. 2021 Apr;13(8):631-647. doi: 10.2217/epi-2020-0409. Epub 2021 Apr 7. Epigenomics. 2021. PMID: 33823614 Review.
Cited by
-
CORN 2.0 - Condition Orientated Regulatory Networks 2.0.Comput Struct Biotechnol J. 2025 Apr 3;27:1518-1528. doi: 10.1016/j.csbj.2025.04.003. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40270708 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

