Psi promotes Drosophila wing growth via direct transcriptional activation of cell cycle targets and repression of growth inhibitors
- PMID: 36692218
- PMCID: PMC10110491
- DOI: 10.1242/dev.201563
Psi promotes Drosophila wing growth via direct transcriptional activation of cell cycle targets and repression of growth inhibitors
Abstract
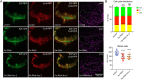
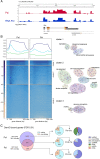
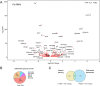
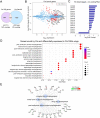
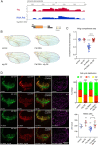
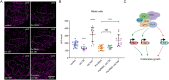
The first characterised FUSE Binding Protein family member, FUBP1, binds single-stranded DNA to activate MYC transcription. Psi, the sole FUBP protein in Drosophila, binds RNA to regulate P-element and mRNA splicing. Our previous work revealed pro-growth functions for Psi, which depend, in part, on transcriptional activation of Myc. Genome-wide functions for FUBP family proteins in transcriptional control remain obscure. Here, through the first genome-wide binding and expression profiles obtained for a FUBP family protein, we demonstrate that, in addition to being required to activate Myc to promote cell growth, Psi also directly binds and activates stg to couple growth and cell division. Thus, Psi knockdown results in reduced cell division in the wing imaginal disc. In addition to activating these pro-proliferative targets, Psi directly represses transcription of the growth inhibitor tolkin (tok, a metallopeptidase implicated in TGFβ signalling). We further demonstrate tok overexpression inhibits proliferation, while tok loss of function increases mitosis alone and suppresses impaired cell division caused by Psi knockdown. Thus, Psi orchestrates growth through concurrent transcriptional activation of the pro-proliferative genes Myc and stg, in combination with repression of the growth inhibitor tok.
Keywords: Cell cycle; Drosophila; FUBP1; Myc; Psi; Transcription.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
Similar articles
-
FUBP/KH domain proteins in transcription: Back to the future.Transcription. 2017 May 27;8(3):185-192. doi: 10.1080/21541264.2017.1293595. Epub 2017 Feb 16. Transcription. 2017. PMID: 28301294 Free PMC article. Review.
-
Transcriptional repression of Myc underlies the tumour suppressor function of AGO1 in Drosophila.Development. 2020 Jun 11;147(11):dev190231. doi: 10.1242/dev.190231. Development. 2020. PMID: 32527935 Free PMC article.
-
Defining the essential function of FBP/KSRP proteins: Drosophila Psi interacts with the mediator complex to modulate MYC transcription and tissue growth.Nucleic Acids Res. 2016 Sep 19;44(16):7646-58. doi: 10.1093/nar/gkw461. Epub 2016 May 20. Nucleic Acids Res. 2016. PMID: 27207882 Free PMC article.
-
Modulo is a target of Myc selectively required for growth of proliferative cells in Drosophila.Mech Dev. 2003 Jun;120(6):645-55. doi: 10.1016/s0925-4773(03)00049-2. Mech Dev. 2003. PMID: 12834864
-
Mechanisms of c-myc-mediated transcriptional repression of growth arrest genes.Exp Cell Res. 2003 Feb 1;283(1):17-21. doi: 10.1016/s0014-4827(02)00020-4. Exp Cell Res. 2003. PMID: 12565816 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

