In-Depth Characterization of Full-Length Archived Viral Genomes after Nine Years of Posttreatment HIV Control
- PMID: 36692300
- PMCID: PMC9927157
- DOI: 10.1128/spectrum.03267-22
In-Depth Characterization of Full-Length Archived Viral Genomes after Nine Years of Posttreatment HIV Control
Abstract
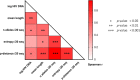
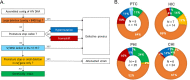
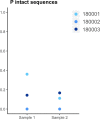
In the search for control of human immunodeficiency virus type 1 (HIV-1) infection without antiretroviral therapy, posttreatment controllers (PTCs) are models of HIV remission. To better understand their mechanisms of control, we characterized the HIV blood reservoirs of 8 PTCs (median of 9.4 years after treatment interruption) in comparison with those of 13 natural HIV infection controllers (HICs) (median of 18 years of infection) and with those of individuals receiving efficient antiretroviral therapy initiated during either primary HIV infection (PHIs; n = 8) or chronic HIV infection (CHIs; n = 6). This characterization was performed with single-genome amplification and deep sequencing. The proviral diversity, which reflects the history of past viral replication, was lower in the PTCs, PHIs, and aviremic HICs than in the blipper HICs and CHIs. The proportions of intact and defective proviruses among the proviral pool in PTCs were not significantly different from those of other groups. When looking at the quantities of proviruses per million peripheral blood mononuclear cells (PBMCs), they had similar amounts of intact proviruses as other groups but smaller amounts of defective proviruses than CHIs, suggesting a role of these forms in HIV pathogenesis. Two HICs but none of the PTCs harbored only proviruses with deletion in nef; these attenuated strains could contribute to viral control in these participants. We show, for the first time, the presence of intact proviruses and low viral diversity in PTCs long after treatment interruption, as well as the absence of evolution of the proviral quasispecies in subsequent samples. This reflects low residual replication over time. Further data are necessary to confirm these results. IMPORTANCE Most people living with HIV need antiretroviral therapy to control their infection and experience viral relapse in case of treatment interruption, because of viral reservoir (proviruses) persistence. Knowing that proviruses are very diverse and most of them are defective in treated individuals, we aimed to characterize the HIV blood reservoirs of posttreatment controllers (PTCs), rare models of drug-free remission, in comparison with spontaneous controllers and treated individuals. At a median time of 9 years after treatment interruption, which is unprecedented in the literature, we showed that the proportions and quantities of intact proviruses were similar between PTCs and other individuals. Unlike 2/7 spontaneous controllers who harbored only nef-deleted proviruses, which are attenuated strains, which could contribute to their control, no such case was observed in PTCs. Furthermore, PTCs displayed low viral genetic diversity and no evolution of their reservoirs, indicating very low residual replication, despite the presence of intact proviruses.
Keywords: HIV reservoirs; defective proviruses; next-generation sequencing; posttreatment HIV controller; posttreatment HIV controllers; provirus; proviruses; ultradeep sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers.J Clin Invest. 2018 Aug 31;128(9):4074-4085. doi: 10.1172/JCI120549. Epub 2018 Aug 20. J Clin Invest. 2018. PMID: 30024859 Free PMC article.
-
HIV Proviral Burden, Genetic Diversity, and Dynamics in Viremic Controllers Who Subsequently Initiated Suppressive Antiretroviral Therapy.mBio. 2021 Dec 21;12(6):e0249021. doi: 10.1128/mBio.02490-21. Epub 2021 Nov 16. mBio. 2021. PMID: 34781741 Free PMC article.
-
Intact HIV Proviruses Persist in Children Seven to Nine Years after Initiation of Antiretroviral Therapy in the First Year of Life.J Virol. 2020 Jan 31;94(4):e01519-19. doi: 10.1128/JVI.01519-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776265 Free PMC article.
-
HIV-1 reservoir landscape of post-treatment control.Curr Opin HIV AIDS. 2025 Jan 1;20(1):99-108. doi: 10.1097/COH.0000000000000891. Epub 2024 Oct 21. Curr Opin HIV AIDS. 2025. PMID: 39484860 Review.
-
"Block and lock" viral integration sites in persons with drug-free control of HIV-1 infection.Curr Opin HIV AIDS. 2024 May 1;19(3):110-115. doi: 10.1097/COH.0000000000000845. Epub 2024 Feb 28. Curr Opin HIV AIDS. 2024. PMID: 38457193 Review.
Cited by
-
Acute HIV-1 Infection: Paradigm and Singularity.Viruses. 2025 Mar 3;17(3):366. doi: 10.3390/v17030366. Viruses. 2025. PMID: 40143294 Free PMC article. Review.
-
Ongoing HIV-1 evolution and reservoir reseeding in two elite controllers with genetically diverse peripheral proviral quasispecies.Mem Inst Oswaldo Cruz. 2023 Jun 5;118:e230066. doi: 10.1590/0074-02760230066. eCollection 2023. Mem Inst Oswaldo Cruz. 2023. PMID: 37283423 Free PMC article.
-
Understanding latent HIV-1 reservoirs through host genomics approaches.iScience. 2023 Oct 28;26(11):108342. doi: 10.1016/j.isci.2023.108342. eCollection 2023 Nov 17. iScience. 2023. PMID: 38026212 Free PMC article. Review.
-
Learning From Full Characterization of HIV Proviruses in People Receiving Long-Acting Cabotegravir/Rilpivirine With a History of Replication on the Antiretroviral Classes.Open Forum Infect Dis. 2024 Dec 24;12(1):ofae748. doi: 10.1093/ofid/ofae748. eCollection 2025 Jan. Open Forum Infect Dis. 2024. PMID: 39872809 Free PMC article.
References
-
- Namazi G, Fajnzylber JM, Aga E, Bosch RJ, Acosta EP, Sharaf R, Hartogensis W, Jacobson JM, Connick E, Volberding P, Skiest D, Margolis D, Sneller MC, Little SJ, Gianella S, Smith DM, Kuritzkes DR, Gulick RM, Mellors JW, Mehraj V, Gandhi RT, Mitsuyasu R, Schooley RT, Henry K, Tebas P, Deeks SG, Chun TW, Collier AC, Routy JP, Hecht FM, Walker BD, Li JZ. 2018. The Control of HIV After Antiretroviral Medication Pause (CHAMP) study: posttreatment controllers identified from 14 clinical studies. J Infect Dis 218:1954–1963. doi:10.1093/infdis/jiy479. - DOI - PMC - PubMed
-
- Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, ANRS VISCONTI Study Group . 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 9:e1003211. doi:10.1371/journal.ppat.1003211. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

