Acetylation of fission yeast tropomyosin does not promote differential association with cognate formins
- PMID: 36692369
- PMCID: PMC10121778
- DOI: 10.1002/cm.21745
Acetylation of fission yeast tropomyosin does not promote differential association with cognate formins
Abstract
It was proposed from cellular studies that S. pombe tropomyosin Cdc8 (Tpm) segregates into two populations due to the presence or absence of an amino-terminal acetylation that specifies which formin-mediated F-actin networks it binds, but with no supporting biochemistry. To address this mechanism in vitro, we developed methods for S. pombe actin expression in Sf9 cells. We then employed 3-color TIRF microscopy using all recombinant S. pombe proteins to probe in vitro multicomponent mechanisms involving actin, acetylated and unacetylated Tpm, formins, and myosins. Acetyl-Tpm exhibits tight binding to actin in contrast to weaker binding by unacetylated Tpm. In disagreement with the differential recruitment model, Tpm showed no preferential binding to filaments assembled by the FH1-FH2-domains of two S. pombe formins, nor did Tpm binding have any bias towards the growing formin-bound actin filament barbed end. Although our in vitro findings do not support a direct formin-tropomyosin interaction, it is possible that formins bias differential tropomyosin isoform recruitment through undiscovered mechanisms. Importantly, despite a 12% sequence divergence between skeletal and S. pombe actin, S. pombe myosins Myo2 and Myo51 exhibited similar motile behavior with these two actins, validating key prior findings with these myosins that used skeletal actin.
Keywords: S. pombe; acetylation; actin; formin; myosin; tropomyosin.
© 2023 The Authors. Cytoskeleton published by Wiley Periodicals LLC.
Conflict of interest statement
Conflict of interest statement
The authors declare no conflict of interest.
Figures
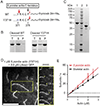




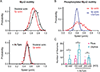

Similar articles
-
Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe.J Cell Sci. 2007 May 1;120(Pt 9):1635-45. doi: 10.1242/jcs.001115. J Cell Sci. 2007. PMID: 17452625
-
Altering the stability of the Cdc8 overlap region modulates the ability of this tropomyosin to bind co-operatively to actin and regulate myosin.Biochem J. 2011 Sep 1;438(2):265-73. doi: 10.1042/BJ20101316. Biochem J. 2011. PMID: 21658004
-
The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast.J Cell Sci. 2010 Oct 1;123(Pt 19):3235-43. doi: 10.1242/jcs.069971. Epub 2010 Aug 31. J Cell Sci. 2010. PMID: 20807799 Free PMC article.
-
Tropomyosin - master regulator of actin filament function in the cytoskeleton.J Cell Sci. 2015 Aug 15;128(16):2965-74. doi: 10.1242/jcs.172502. Epub 2015 Aug 3. J Cell Sci. 2015. PMID: 26240174 Review.
-
Coordination of microtubule acetylation and the actin cytoskeleton by formins.Cell Mol Life Sci. 2018 Sep;75(17):3181-3191. doi: 10.1007/s00018-018-2855-3. Epub 2018 Jun 15. Cell Mol Life Sci. 2018. PMID: 29947928 Free PMC article. Review.
Cited by
-
Functional redundancy and formin-independent localization of tropomyosin isoforms in Saccharomyces cerevisiae.bioRxiv [Preprint]. 2024 Dec 11:2024.04.04.587703. doi: 10.1101/2024.04.04.587703. bioRxiv. 2024. PMID: 38617342 Free PMC article. Preprint.
-
Crystal structures of cables formed by the acetylated and unacetylated forms of the Schizosaccharomyces pombe tropomyosin ortholog TpmCdc8.J Biol Chem. 2024 Dec;300(12):107925. doi: 10.1016/j.jbc.2024.107925. Epub 2024 Oct 25. J Biol Chem. 2024. PMID: 39461476 Free PMC article.
References
-
- Balasubramanian MK, Helfman DM, and Hemmingsen SM (1992). A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature 360, 84–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

