Selective integration of diverse taste inputs within a single taste modality
- PMID: 36692370
- PMCID: PMC9873257
- DOI: 10.7554/eLife.84856
Selective integration of diverse taste inputs within a single taste modality
Abstract
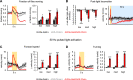
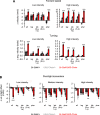
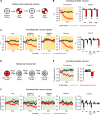
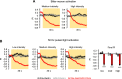
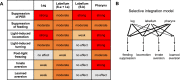
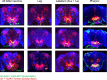
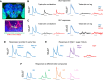
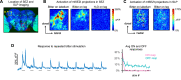
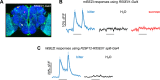
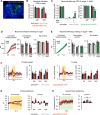
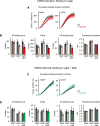
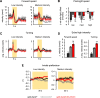
A fundamental question in sensory processing is how different channels of sensory input are processed to regulate behavior. Different input channels may converge onto common downstream pathways to drive the same behaviors, or they may activate separate pathways to regulate distinct behaviors. We investigated this question in the Drosophila bitter taste system, which contains diverse bitter-sensing cells residing in different taste organs. First, we optogenetically activated subsets of bitter neurons within each organ. These subsets elicited broad and highly overlapping behavioral effects, suggesting that they converge onto common downstream pathways, but we also observed behavioral differences that argue for biased convergence. Consistent with these results, transsynaptic tracing revealed that bitter neurons in different organs connect to overlapping downstream pathways with biased connectivity. We investigated taste processing in one type of downstream bitter neuron that projects to the higher brain. These neurons integrate input from multiple organs and regulate specific taste-related behaviors. We then traced downstream circuits, providing the first glimpse into taste processing in the higher brain. Together, these results reveal that different bitter inputs are selectively integrated early in the circuit, enabling the pooling of information, while the circuit then diverges into multiple pathways that may have different roles.
Keywords: D. melanogaster; feeding; neural circuits; neuroscience; sensory processing; taste.
© 2023, Deere et al.
Conflict of interest statement
JD, AS, MY, HU, NM, LN, KR, AD No competing interests declared
Figures



Similar articles
-
Connectomic analysis of taste circuits in Drosophila.Sci Rep. 2025 Feb 12;15(1):5278. doi: 10.1038/s41598-025-89088-9. Sci Rep. 2025. PMID: 39939650 Free PMC article.
-
Complex representation of taste quality by second-order gustatory neurons in Drosophila.Curr Biol. 2022 Sep 12;32(17):3758-3772.e4. doi: 10.1016/j.cub.2022.07.048. Epub 2022 Aug 15. Curr Biol. 2022. PMID: 35973432 Free PMC article.
-
Behavioral Analysis of Bitter Taste Perception in Drosophila Larvae.Chem Senses. 2016 Jan;41(1):85-94. doi: 10.1093/chemse/bjv061. Epub 2015 Oct 28. Chem Senses. 2016. PMID: 26512069
-
Regulation of feeding behavior in Drosophila through the interplay of gustation, physiology and neuromodulation.Front Biosci (Landmark Ed). 2018 Jun 1;23(11):2016-2027. doi: 10.2741/4686. Front Biosci (Landmark Ed). 2018. PMID: 29772542 Review.
-
Gustatory processing and taste memory in Drosophila.J Neurogenet. 2016 Jun;30(2):112-21. doi: 10.1080/01677063.2016.1185104. J Neurogenet. 2016. PMID: 27328844 Free PMC article. Review.
Cited by
-
Taste cues elicit prolonged modulation of feeding behavior in Drosophila.iScience. 2022 Sep 17;25(10):105159. doi: 10.1016/j.isci.2022.105159. eCollection 2022 Oct 21. iScience. 2022. PMID: 36204264 Free PMC article.
-
Connectomic analysis of taste circuits in Drosophila.Sci Rep. 2025 Feb 12;15(1):5278. doi: 10.1038/s41598-025-89088-9. Sci Rep. 2025. PMID: 39939650 Free PMC article.
-
Connectomic analysis of taste circuits in Drosophila.bioRxiv [Preprint]. 2024 Sep 15:2024.09.14.613080. doi: 10.1101/2024.09.14.613080. bioRxiv. 2024. Update in: Sci Rep. 2025 Feb 12;15(1):5278. doi: 10.1038/s41598-025-89088-9. PMID: 39314399 Free PMC article. Updated. Preprint.
-
Driver lines for studying associative learning in Drosophila.Elife. 2025 Jan 29;13:RP94168. doi: 10.7554/eLife.94168. Elife. 2025. PMID: 39879130 Free PMC article.
-
Hierarchical diversification of instinctual behavior neurons by lineage, birth order, and sex.bioRxiv [Preprint]. 2025 Jun 3:2025.06.03.657692. doi: 10.1101/2025.06.03.657692. bioRxiv. 2025. PMID: 40502082 Free PMC article. Preprint.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

