METTL3 from Target Validation to the First Small-Molecule Inhibitors: A Medicinal Chemistry Journey
- PMID: 36692498
- PMCID: PMC9923689
- DOI: 10.1021/acs.jmedchem.2c01601
METTL3 from Target Validation to the First Small-Molecule Inhibitors: A Medicinal Chemistry Journey
Abstract
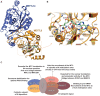
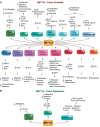
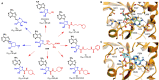
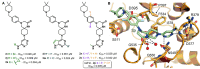
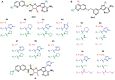
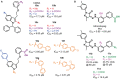
RNA methylation is a critical mechanism for regulating the transcription and translation of specific sequences or for eliminating unnecessary sequences during RNA maturation. METTL3, an RNA methyltransferase that catalyzes the transfer of a methyl group to the N6-adenosine of RNA, is one of the key mediators of this process. METTL3 dysregulation may result in the emergence of a variety of diseases ranging from cancer to cardiovascular and neurological disorders beyond contributing to viral infections. Hence, the discovery of METTL3 inhibitors may assist in furthering the understanding of the biological roles of this enzyme, in addition to contributing to the development of novel therapeutics. Through this work, we will examine the existing correlations between METTL3 and diseases. We will also analyze the development, mode of action, pharmacology, and structure-activity relationships of the currently known METTL3 inhibitors. They include both nucleoside and non-nucleoside compounds, with the latter comprising both competitive and allosteric inhibitors.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
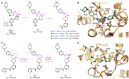
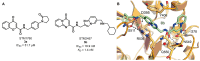

Similar articles
-
Discovery of substituted indole derivatives as allosteric inhibitors of m6 A-RNA methyltransferase, METTL3-14 complex.Drug Dev Res. 2022 May;83(3):783-799. doi: 10.1002/ddr.21910. Epub 2022 Jan 18. Drug Dev Res. 2022. PMID: 35040501
-
RNA secondary structure dependence in METTL3-METTL14 mRNA methylation is modulated by the N-terminal domain of METTL3.Biol Chem. 2020 Oct 19;402(1):89-98. doi: 10.1515/hsz-2020-0265. Print 2020 Nov 18. Biol Chem. 2020. PMID: 33544495
-
Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex.Nature. 2016 Jun 23;534(7608):575-8. doi: 10.1038/nature18298. Epub 2016 May 25. Nature. 2016. PMID: 27281194
-
Roles of METTL3 in cancer: mechanisms and therapeutic targeting.J Hematol Oncol. 2020 Aug 27;13(1):117. doi: 10.1186/s13045-020-00951-w. J Hematol Oncol. 2020. PMID: 32854717 Free PMC article. Review.
-
METTL3 plays a crucial function in multiple biological processes.Acta Histochem. 2022 Aug;124(6):151916. doi: 10.1016/j.acthis.2022.151916. Epub 2022 Jun 22. Acta Histochem. 2022. PMID: 35752056 Review.
Cited by
-
Virtual Screening and Molecular Docking: Discovering Novel METTL3 Inhibitors.ACS Med Chem Lett. 2024 Aug 6;15(9):1491-1499. doi: 10.1021/acsmedchemlett.4c00216. eCollection 2024 Sep 12. ACS Med Chem Lett. 2024. PMID: 39291017
-
METTL3: a multifunctional regulator in diseases.Mol Cell Biochem. 2025 Jun;480(6):3429-3454. doi: 10.1007/s11010-025-05208-z. Epub 2025 Jan 24. Mol Cell Biochem. 2025. PMID: 39853661 Review.
-
Patent landscape of small molecule inhibitors of METTL3 (2020-present).Expert Opin Ther Pat. 2024 Dec 26:1-16. doi: 10.1080/13543776.2024.2447056. Online ahead of print. Expert Opin Ther Pat. 2024. PMID: 39721070 Review.
-
Nurturing gut health: role of m6A RNA methylation in upholding the intestinal barrier.Cell Death Discov. 2024 Jun 3;10(1):271. doi: 10.1038/s41420-024-02043-x. Cell Death Discov. 2024. PMID: 38830900 Free PMC article. Review.
-
SRF Facilitates Transcriptional Inhibition of Gem Expression by m6A Methyltransferase METTL3 to Suppress Neuronal Damage in Epilepsy.Mol Neurobiol. 2025 Mar;62(3):2903-2925. doi: 10.1007/s12035-024-04396-x. Epub 2024 Aug 27. Mol Neurobiol. 2025. PMID: 39190265
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

