METTL3 is essential for normal progesterone signaling during embryo implantation via m6A-mediated translation control of progesterone receptor
- PMID: 36693099
- PMCID: PMC9945998
- DOI: 10.1073/pnas.2214684120
METTL3 is essential for normal progesterone signaling during embryo implantation via m6A-mediated translation control of progesterone receptor
Abstract
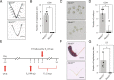
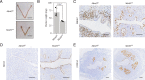
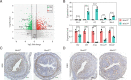
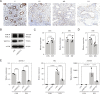
Embryo implantation, a crucial step in human reproduction, is tightly controlled by estrogen and progesterone (P4) via estrogen receptor alpha and progesterone receptor (PGR), respectively. Here, we report that N6-methyladenosine (m6A), the most abundant mRNA modification in eukaryotes, plays an essential role in embryo implantation through the maintenance of P4 signaling. Conditional deletion of methyltransferase-like 3 (Mettl3), encoding the m6A writer METTL3, in the female reproductive tract using a Cre mouse line with Pgr promoter (Pgr-Cre) resulted in complete implantation failure due to pre-implantation embryo loss and defective uterine receptivity. Moreover, the uterus of Mettl3 null mice failed to respond to artificial decidualization. We further found that Mettl3 deletion was accompanied by a marked decrease in PGR protein expression. Mechanistically, we found that Pgr mRNA is a direct target for METTL3-mediated m6A modification. A luciferase assay revealed that the m6A modification in the 5' untranslated region (5'-UTR) of Pgr mRNA enhances PGR protein translation efficiency in a YTHDF1-dependent manner. Finally, we demonstrated that METTL3 is required for human endometrial stromal cell decidualization in vitro and that the METTL3-PGR axis is conserved between mice and humans. In summary, this study provides evidence that METTL3 is essential for normal P4 signaling during embryo implantation via m6A-mediated translation control of Pgr mRNA.
Keywords: METTL3; embryo implantation; m6A; progesterone receptor.
Conflict of interest statement
The authors declare no competing interest.
Figures





Similar articles
-
METTL3-dependent m6A methylation facilitates uterine receptivity and female fertility via balancing estrogen and progesterone signaling.Cell Death Dis. 2023 Jun 3;14(6):349. doi: 10.1038/s41419-023-05866-1. Cell Death Dis. 2023. PMID: 37270544 Free PMC article.
-
Uterine Epithelial Progesterone Receptor Governs Uterine Receptivity Through Epithelial Cell Differentiation.Endocrinology. 2020 Dec 1;161(12):bqaa195. doi: 10.1210/endocr/bqaa195. Endocrinology. 2020. PMID: 33099617
-
The METTL3-PGR-WNT4 axis is critical for human endometrial stromal cell decidualization.FEBS J. 2025 Jul 25. doi: 10.1111/febs.70200. Online ahead of print. FEBS J. 2025. PMID: 40711993
-
In vivo analysis of progesterone receptor action in the uterus during embryo implantation.Semin Cell Dev Biol. 2008 Apr;19(2):178-86. doi: 10.1016/j.semcdb.2007.12.001. Epub 2008 Jan 6. Semin Cell Dev Biol. 2008. PMID: 18280760 Review.
-
Progress on the Role of Estrogen and Progesterone Signaling in Mouse Embryo Implantation and Decidualization.Reprod Sci. 2023 Jun;30(6):1746-1757. doi: 10.1007/s43032-023-01169-0. Epub 2023 Jan 24. Reprod Sci. 2023. PMID: 36694081 Review.
Cited by
-
Structural proteomics defines a sequential priming mechanism for the progesterone receptor.Nat Commun. 2025 May 12;16(1):4403. doi: 10.1038/s41467-025-59458-y. Nat Commun. 2025. PMID: 40355435 Free PMC article.
-
Emerging roles of RNA-binding proteins in fatty liver disease.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1840. doi: 10.1002/wrna.1840. Wiley Interdiscip Rev RNA. 2024. PMID: 38613185 Free PMC article. Review.
-
Mettl3/Eed/Ythdc1 regulatory axis controls endometrial receptivity and function.Commun Biol. 2025 Feb 11;8(1):215. doi: 10.1038/s42003-025-07667-y. Commun Biol. 2025. PMID: 39934221 Free PMC article.
-
Structural proteomics defines a sequential priming mechanism for the progesterone receptor.Res Sq [Preprint]. 2024 Nov 14:rs.3.rs-5199635. doi: 10.21203/rs.3.rs-5199635/v1. Res Sq. 2024. Update in: Nat Commun. 2025 May 12;16(1):4403. doi: 10.1038/s41467-025-59458-y. PMID: 39606477 Free PMC article. Updated. Preprint.
-
IGF2BP2 regulates the proliferation and migration of endometrial stromal cells through the PI3K/AKT/mTOR signaling pathway in Hu sheep.J Anim Sci. 2024 Jan 3;102:skae129. doi: 10.1093/jas/skae129. J Anim Sci. 2024. PMID: 38727196 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

