Gene targeting in amyotrophic lateral sclerosis using causality-based feature selection and machine learning
- PMID: 36694130
- PMCID: PMC9872307
- DOI: 10.1186/s10020-023-00603-y
Gene targeting in amyotrophic lateral sclerosis using causality-based feature selection and machine learning
Abstract
Background: Amyotrophic lateral sclerosis (ALS) is a rare progressive neurodegenerative disease that affects upper and lower motor neurons. As the molecular basis of the disease is still elusive, the development of high-throughput sequencing technologies, combined with data mining techniques and machine learning methods, could provide remarkable results in identifying pathogenetic mechanisms. High dimensionality is a major problem when applying machine learning techniques in biomedical data analysis, since a huge number of features is available for a limited number of samples. The aim of this study was to develop a methodology for training interpretable machine learning models in the classification of ALS and ALS-subtypes samples, using gene expression datasets.
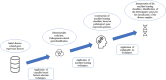
Methods: We performed dimensionality reduction in gene expression data using a semi-automated preprocessing systematic gene selection procedure using Statistically Equivalent Signature (SES), a causality-based feature selection algorithm, followed by Boosted Regression Trees (XGBoost) and Random Forest to train the machine learning classifiers. The SHapley Additive exPlanations (SHAP values) were used for interpretation of the machine learning classifiers. The methodology was developed and tested using two distinct publicly available ALS RNA-seq datasets. We evaluated the performance of SES as a dimensionality reduction method against: (a) Least Absolute Shrinkage and Selection Operator (LASSO), and (b) Local Outlier Factor (LOF).
Results: The proposed methodology achieved 85.18% accuracy for the classification of cerebellum or frontal cortex samples as C9orf72-related familial ALS, sporadic ALS or healthy samples. Importantly, the genes identified as the most determinative have also been reported as disease-associated in ALS literature. When tested in the evaluation dataset, the methodology achieved 88.89% accuracy for the classification of sporadic ALS motor neuron samples. When LASSO was used as feature selection method instead of SES, the accuracy of the machine learning classifiers ranged from 74.07 to 96.30%, depending on tissue assessed, while LOF underperformed significantly (77.78% accuracy for the classification of pooled cerebellum and frontal cortex samples).
Conclusions: Using SES, we addressed the challenge of high dimensionality in gene expression data analysis, and we trained accurate machine learning ALS classifiers, specific for the gene expression patterns of different disease subtypes and tissue samples, while identifying disease-associated genes.
Keywords: Causality-based feature selection; Dimensionality reduction; Gene expression; Machine learning.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Molecular Classification and Interpretation of Amyotrophic Lateral Sclerosis Using Deep Convolution Neural Networks and Shapley Values.Genes (Basel). 2021 Oct 30;12(11):1754. doi: 10.3390/genes12111754. Genes (Basel). 2021. PMID: 34828360 Free PMC article.
-
Model-Based and Model-Free Techniques for Amyotrophic Lateral Sclerosis Diagnostic Prediction and Patient Clustering.Neuroinformatics. 2019 Jul;17(3):407-421. doi: 10.1007/s12021-018-9406-9. Neuroinformatics. 2019. PMID: 30460455 Free PMC article.
-
Unsupervised machine learning identifies distinct ALS molecular subtypes in post-mortem motor cortex and blood expression data.Acta Neuropathol Commun. 2023 Dec 21;11(1):208. doi: 10.1186/s40478-023-01686-8. Acta Neuropathol Commun. 2023. PMID: 38129934 Free PMC article.
-
Feature selection from magnetic resonance imaging data in ALS: a systematic review.Ther Adv Chronic Dis. 2021 Oct 13;12:20406223211051002. doi: 10.1177/20406223211051002. eCollection 2021. Ther Adv Chronic Dis. 2021. PMID: 34729157 Free PMC article. Review.
-
What Can Machine Learning Approaches in Genomics Tell Us about the Molecular Basis of Amyotrophic Lateral Sclerosis?J Pers Med. 2020 Nov 26;10(4):247. doi: 10.3390/jpm10040247. J Pers Med. 2020. PMID: 33256133 Free PMC article. Review.
Cited by
-
Machine learning in rare disease.Nat Methods. 2023 Jun;20(6):803-814. doi: 10.1038/s41592-023-01886-z. Epub 2023 May 29. Nat Methods. 2023. PMID: 37248386 Review.
-
Role and Potential of Artificial Intelligence in Biomarker Discovery and Development of Treatment Strategies for Amyotrophic Lateral Sclerosis.Int J Mol Sci. 2025 May 2;26(9):4346. doi: 10.3390/ijms26094346. Int J Mol Sci. 2025. PMID: 40362582 Free PMC article. Review.
-
The Effect of Naturally Acquired Immunity on Mortality Predictors: A Focus on Individuals with New Coronavirus.Biomedicines. 2025 Mar 27;13(4):803. doi: 10.3390/biomedicines13040803. Biomedicines. 2025. PMID: 40299374 Free PMC article.
-
Exploring the role of candidalysin in the pathogenicity of Candida albicans by gene set enrichment analysis and evolutionary dynamics.Am J Transl Res. 2024 Jul 15;16(7):3191-3210. doi: 10.62347/IZYM9087. eCollection 2024. Am J Transl Res. 2024. PMID: 39114682 Free PMC article.
-
Leveraging machine learning for precision medicine: a predictive model for cognitive impairment in cholestasis patients.BMC Gastroenterol. 2025 Mar 18;25(1):185. doi: 10.1186/s12876-025-03711-7. BMC Gastroenterol. 2025. PMID: 40102737 Free PMC article.
References
-
- Anna Roumpelaki KB. Package “MXM” Type Package Title Feature Selection (Including Multiple Solutions) and Bayesian Networks. 2022. https://cran.r-project.org/web/packages/MXM/MXM.pdf
-
- Barredo Arrieta A, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion. 2020;1(58):82–115. doi: 10.1016/j.inffus.2019.12.012. - DOI
-
- Batra R, Hutt K, Vu A, Rabin SJ, Baughn MW, Libby RT, et al. Gene Expression Signatures of Sporadic ALS Motor Neuron Populations. Neuroscience. 2016 doi: 10.1101/038448. - DOI
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

