Goat FADS2 controlling fatty acid metabolism is directly regulated by SREBP1 in mammary epithelial cells
- PMID: 36694375
- PMCID: PMC9982361
- DOI: 10.1093/jas/skad030
Goat FADS2 controlling fatty acid metabolism is directly regulated by SREBP1 in mammary epithelial cells
Abstract
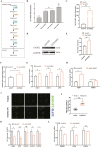
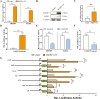
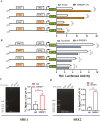
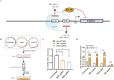
Goat milk provides benefits to human health due to its richness in bioactive components, such as polyunsaturated fatty acids (PUFAs). The fatty acid desaturase 2 (FADS2) is the first rate-limiting enzyme in PUFAs biosynthesis. However, its role and transcriptional regulation mechanisms in fatty acid metabolism in dairy goat remain unclear. Here, our study revealed that the FADS2 gene was highly expressed during the peak lactation compared with the dry period, early lactation, and late lactation. The content of triacylglycerol (TAG) was enhanced with the increasing mRNA expression of TAG synthesis genes (diacylglycerol acyltransferase 1/2, DGAT1/2) in FADS2-overexpressed goat mammary epithelial cells (GMECs). Overexpression of FADS2 was positively correlated with the elevated concentrations of dihomo-gamma-linolenic acid (DGLA) and docosahexaenoic acid (DHA) in GMECs. BODIPY staining showed that FADS2 promoted lipid droplet accumulation in GMECs. To clarify the transcriptional regulatory mechanisms of FADS2, 2,226 bp length of FADS2 promoter was obtained. Deletion mutation assays revealed that the core region of FADS2 promoter was located between the -375 and -26 region, which contained SRE1 (-361/-351) and SRE2 (-191/-181) cis-acting elements of transcription factor sterol regulatory element-binding protein 1 (SREBP1). Overexpression of SREBP1 enhanced relative luciferase activity of the single mutant of SRE1 or SRE2, vice versa, and failed to alter the relative luciferase activity of the joint mutant of SRE1 and SRE2. Chromatin immunoprecipitation (ChIP) and site-directed mutation assays further demonstrated that SREBP1 regulated the transcription of the FADS2 gene by binding to SRE sites in vivo and in vitro. In addition, the mRNA levels of FADS2 were significantly decreased by targeting SRE1 and SRE2 sites in the genome via the CRISPR interference (CRISPRi) system. These findings establish a direct role for FADS2 regulating TAG and fatty acid synthesis by SREBP1 transcriptional regulation in dairy goat, providing new insights into fatty acid metabolism in mammary gland of ruminants.
Keywords: FADS2; SREBP1; GMECs; fatty acid synthesis; transcriptional regulation.
Plain language summary
The fatty acid desaturase 2 (FADS2) is the first rate-limiting enzyme in polyunsaturated fatty acids (PUFAs) biosynthesis in mammals. This study aimed to investigate the function and transcriptional regulation mechanism of FADS2 in goat mammary epithelial cells (GMECs). The content of triacylglycerol (TAG) was enhanced with lipid droplet accumulation in FADS2-overexpressed GMECs. Overexpression of FADS2 was positively correlated with elevated concentrations of docosahexaenoic acid (DHA) in GMECs. Furthermore, site-directed mutation and chromatin immunoprecipitation (ChIP) assays simultaneously demonstrated that FADS2 was directly regulated by SREBP1 transcriptional factor binding to sterol regulatory element (SRE) in vitro and in vivo. In addition, genetic ablation of SRE1 and SRE2 in the genome resulted in a significant reduction in the mRNA levels of FADS2 via the CRISPR interference (CRISPRi) system. Altogether, this study discovered that the SREBP1 exerts control on FADS2 to regulate milk fatty acids, and provides a theoretical approach for improving milk quality via genetic approaches.
© The Author(s) 2023. Published by Oxford University Press on behalf of the American Society of Animal Science. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
C/EBPα promotes triacylglycerol synthesis via regulating PPARG promoter activity in goat mammary epithelial cells.J Anim Sci. 2023 Jan 3;101:skac412. doi: 10.1093/jas/skac412. J Anim Sci. 2023. PMID: 36547378 Free PMC article.
-
The FoxO1-ATGL axis alters milk lipolysis homeostasis through PI3K/AKT signaling pathway in dairy goat mammary epithelial cells.J Anim Sci. 2023 Jan 3;101:skad286. doi: 10.1093/jas/skad286. J Anim Sci. 2023. PMID: 37638641 Free PMC article.
-
FoxO1 Knockdown Promotes Fatty Acid Synthesis via Modulating SREBP1 Activities in the Dairy Goat Mammary Epithelial Cells.J Agric Food Chem. 2020 Oct 28;68(43):12067-12078. doi: 10.1021/acs.jafc.0c05237. Epub 2020 Oct 15. J Agric Food Chem. 2020. PMID: 33054209
-
Effect of short-chain fatty acids on triacylglycerol accumulation, lipid droplet formation and lipogenic gene expression in goat mammary epithelial cells.Anim Sci J. 2016 Feb;87(2):242-9. doi: 10.1111/asj.12420. Epub 2015 Aug 24. Anim Sci J. 2016. PMID: 26304676 Review.
-
An allostatic control of membrane lipid composition by SREBP1.FEBS Lett. 2010 Jun 18;584(12):2689-98. doi: 10.1016/j.febslet.2010.04.004. Epub 2010 Apr 10. FEBS Lett. 2010. PMID: 20385130 Review.
Cited by
-
Genetic Variants Affecting FADS2 Enzyme Dynamics and Gene Expression in Cogenetic Oysters with Different PUFA Levels Provide New Tools to Improve Unsaturated Fatty Acids.Int J Mol Sci. 2024 Dec 18;25(24):13551. doi: 10.3390/ijms252413551. Int J Mol Sci. 2024. PMID: 39769316 Free PMC article.
-
Transcriptomic Profiling Reveals Lysine-Mediated Proliferative Mechanisms in Mongolian Horse Myogenic Satellite Cells.Animals (Basel). 2025 Jun 9;15(12):1711. doi: 10.3390/ani15121711. Animals (Basel). 2025. PMID: 40564262 Free PMC article.
References
-
- Ashikawa, Y., Nishimura Y., Okabe S., Sato Y., Yuge M., Tada T., Miyao H., Murakami S., Kawaguchi K., Sasagawa S.. et al. 2017. Potential protective function of the sterol regulatory element binding factor 1-fatty acid desaturase 1/2 axis in early-stage age-related macular degeneration. Heliyon. 3:e00266. doi:10.1016/j.heliyon.2017.e00266 - DOI - PMC - PubMed
-
- Datsomor, A. K., Olsen R. E., Zic N., Madaro A., Bones A. M., Edvardsen R. B., Wargelius A., and Winge P.. . 2019. CRISPR/Cas9-mediated editing of Delta5 and Delta6 desaturases impairs Delta8-desaturation and docosahexaenoic acid synthesis in Atlantic salmon (Salmo salar L.). Sci. Rep. 9:16888. doi:10.1038/s41598-019-53316-w - DOI - PMC - PubMed
-
- Delgadillo-Puga, C., and Cuchillo-Hilario M.. . 2021. Reviewing the benefits of grazing/browsing semiarid rangeland feed resources and the transference of bioactivity and pro-healthy properties to goat milk and cheese: obesity, insulin resistance, inflammation and hepatic steatosis prevention. Animals. 11:2942. doi:10.3390/ani11102942 - DOI - PMC - PubMed
-
- Delgadillo-Puga, C., Noriega L. G., Morales-Romero A. M., Nieto-Camacho A., Granados-Portillo O., Rodríguez-López L. A., Alemán G., Furuzawa-Carballeda J., Tovar A. R., Cisneros-Zevallos L.. et al. 2020. Goat’s milk intake prevents obesity, hepatic steatosis and insulin resistance in mice fed a high-fat diet by reducing inflammatory markers and increasing energy expenditure and mitochondrial content in skeletal muscle. Int. J. Mol. Sci. 21:5530. doi:10.3390/ijms21155530 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

