circ-IARS depletion inhibits the progression of non-small-cell lung cancer by circ-IARS/miR-1252-5p/HDGF ceRNA pathway
- PMID: 36694627
- PMCID: PMC9835196
- DOI: 10.1515/med-2022-0613
circ-IARS depletion inhibits the progression of non-small-cell lung cancer by circ-IARS/miR-1252-5p/HDGF ceRNA pathway
Abstract
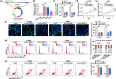
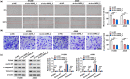
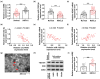
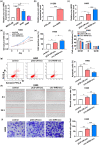
This study aims to explore the role and mechanism of circ-IARS in non-small-cell lung cancer (NSCLC) progression. Expression of circ-IARS, microRNA (miR)-1252-5p, and hepatoma-derived growth factor (HDGF) was measured by real-time quantitative PCR and western blotting. The interactions among circ-IARS, miR-1252-5p, and HDGF were determined by dual-luciferase reporter assay and RNA immunoprecipitation. Cell behaviors were measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 5-ethynyl-2'-deoxyuridine (EdU) assay, flow cytometry, scratch wound assay, and transwell assay, and validated in in vivo xenograft model. Exosomes were isolated using commercial kit, and the expression and functions of exosomal circ-IARS (exo-circ-IARS) were analyzed as described above. Results showed that the expression of circ-IARS was upregulated in NSCLC cells, NSCLC tissues, and serum exosomes from NSCLC patients. circ-IARS exhaustion antagonized cell proliferation, cell cycle progression, migration, and invasion and promoted apoptosis in NSCLC. Molecularly, circ-IARS could sponge miR-1252-5p to modulate the expression of the downstream gene HDGF. In addition, miR-1252-5p downregulation attenuated circ-IARS exhaustion-mediated effects in H1299 and A549 cells. MiR-1252-5p mimic-induced effects were relieved by increasing HDGF expression in H1299 and A549 cells. Exo-circ-IARS promoted H460 cell proliferation, migration, and invasion and inhibited cell apoptosis. Silencing circ-IARS retarded tumor growth of NSCLC cells in vivo. Thus, circ-IARS, secreted by exosomes, was a novel oncogene in NSCLC and regulated the malignant development of NSCLC cells via circ-IARS/miR-1252-5p/HDGF competing endogenous RNA regulatory axis.
Keywords: HDGF; NSCLC; circ-IARS; exosome; miR-1252-5p.
© 2023 the author(s), published by De Gruyter.
Conflict of interest statement
Conflict of interest: The authors declare that they have no competing interest.
Figures





Similar articles
-
Downregulation of circ_PLXND1 inhibits tumorigenesis of non-small cell lung carcinoma via miR-1287-5p/ERBB3 axis.Thorac Cancer. 2023 Jun;14(17):1543-1555. doi: 10.1111/1759-7714.14897. Epub 2023 Apr 18. Thorac Cancer. 2023. PMID: 37073425 Free PMC article.
-
Circ_SETD3 regulates gefitinib sensitivity and tumor progression by miR-873-5p-dependent regulation of APPBP2 in non-small cell lung cancer.J Chemother. 2022 Oct;34(6):401-413. doi: 10.1080/1120009X.2021.2009991. Epub 2021 Dec 3. J Chemother. 2022. PMID: 34861803
-
Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis.Drug Deliv. 2022 Dec;29(1):1257-1271. doi: 10.1080/10717544.2022.2057617. Drug Deliv. 2022. PMID: 35467477 Free PMC article.
-
Hsa_circ_0017639 regulates cisplatin resistance and tumor growth via acting as a miR-1296-5p molecular sponge and modulating sine oculis homeobox 1 expression in non-small cell lung cancer.Bioengineered. 2022 Apr;13(4):8806-8822. doi: 10.1080/21655979.2022.2053810. Bioengineered. 2022. PMID: 35287543 Free PMC article.
-
circ_0007385 served as competing endogenous RNA for miR-519d-3p to suppress malignant behaviors and cisplatin resistance of non-small cell lung cancer cells.Thorac Cancer. 2020 Aug;11(8):2196-2208. doi: 10.1111/1759-7714.13527. Epub 2020 Jun 29. Thorac Cancer. 2020. PMID: 32602212 Free PMC article.
Cited by
-
Non-coding RNAs and exosomal non-coding RNAs in lung cancer: insights into their functions.Front Cell Dev Biol. 2024 May 27;12:1397788. doi: 10.3389/fcell.2024.1397788. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38859962 Free PMC article. Review.
-
Circular RNAs in human diseases.MedComm (2020). 2024 Sep 4;5(9):e699. doi: 10.1002/mco2.699. eCollection 2024 Sep. MedComm (2020). 2024. PMID: 39239069 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. - PubMed
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. - PubMed
-
- Mesko S, Gomez D. Proton therapy in non-small cell lung cancer. Curr Treat Options Oncol. 2018;19(12):76. - PubMed
LinkOut - more resources
Full Text Sources
