Flow-Seq Method: Features and Application in Bacterial Translation Studies
- PMID: 36694903
- PMCID: PMC9844084
- DOI: 10.32607/actanaturae.11820
Flow-Seq Method: Features and Application in Bacterial Translation Studies
Abstract
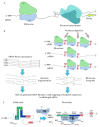
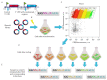
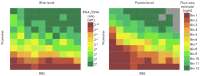
The Flow-seq method is based on using reporter construct libraries, where a certain element regulating the gene expression of fluorescent reporter proteins is represented in many thousands of variants. Reporter construct libraries are introduced into cells, sorted according to their fluorescence level, and then subjected to next-generation sequencing. Therefore, it turns out to be possible to identify patterns that determine the expression efficiency, based on tens and hundreds of thousands of reporter constructs in one experiment. This method has become common in evaluating the efficiency of protein synthesis simultaneously by multiple mRNA variants. However, its potential is not confined to this area. In the presented review, a comparative analysis of the Flow-seq method and other alternative approaches used for translation efficiency evaluation of mRNA was carried out; the features of its application and the results obtained by Flow-seq were also considered.
Keywords: Flow-seq; NGS; bacteria; flow cytometry; high-throughput sequencing; translation.
Copyright ® 2022 National Research University Higher School of Economics.
Figures

Similar articles
-
Flow-Seq Evaluation of Translation Driven by a Set of Natural Escherichia coli 5'-UTR of Variable Length.Int J Mol Sci. 2022 Oct 14;23(20):12293. doi: 10.3390/ijms232012293. Int J Mol Sci. 2022. PMID: 36293163 Free PMC article.
-
Application of sorting and next generation sequencing to study 5΄-UTR influence on translation efficiency in Escherichia coli.Nucleic Acids Res. 2017 Apr 7;45(6):3487-3502. doi: 10.1093/nar/gkw1141. Nucleic Acids Res. 2017. PMID: 27899632 Free PMC article.
-
Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing.Genome Res. 2013 Jun;23(6):977-87. doi: 10.1101/gr.150342.112. Epub 2013 Apr 11. Genome Res. 2013. PMID: 23580730 Free PMC article.
-
Ribosomal profiling adds new coding sequences to the proteome.Biochem Soc Trans. 2015 Dec;43(6):1271-6. doi: 10.1042/BST20150170. Biochem Soc Trans. 2015. PMID: 26614672 Review.
-
Library construction for next-generation sequencing: overviews and challenges.Biotechniques. 2014 Feb 1;56(2):61-4, 66, 68, passim. doi: 10.2144/000114133. eCollection 2014. Biotechniques. 2014. PMID: 24502796 Free PMC article. Review.
Cited by
-
Higher throughput assays for understanding the pathogenicity of variants of unknown significance (VUS) in the RPE65 gene.bioRxiv [Preprint]. 2025 Feb 5:2025.01.31.635952. doi: 10.1101/2025.01.31.635952. bioRxiv. 2025. PMID: 39975398 Free PMC article. Preprint.
References
-
- Shine J., Dalgarno L.. Nature. 1975;254:34–38. - PubMed
LinkOut - more resources
Full Text Sources
