Lamin A/C phosphorylation at serine 22 is a conserved heat shock response to regulate nuclear adaptation during stress
- PMID: 36695453
- PMCID: PMC10022683
- DOI: 10.1242/jcs.259788
Lamin A/C phosphorylation at serine 22 is a conserved heat shock response to regulate nuclear adaptation during stress
Abstract
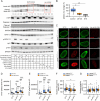
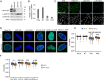
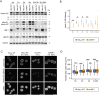
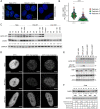
The heat shock (HS) response is crucial for cell survival in harmful environments. Nuclear lamin A/C, encoded by the LMNA gene, contributes towards altered gene expression during HS, but the underlying mechanisms are poorly understood. Here, we show that upon HS, lamin A/C was reversibly phosphorylated at serine 22 in concert with HSF1 activation in human cells, mouse cells and Drosophila melanogaster in vivo. Consequently, the phosphorylation facilitated nucleoplasmic localization of lamin A/C and nuclear sphericity in response to HS. Interestingly, lamin A/C knock-out cells showed deformed nuclei after HS and were rescued by ectopic expression of wild-type lamin A, but not by a phosphomimetic (S22D) lamin A mutant. Furthermore, HS triggered concurrent downregulation of lamina-associated protein 2α (Lap2α, encoded by TMPO) in wild-type lamin A/C-expressing cells, but a similar response was perturbed in lamin A/C knock-out cells and in LMNA mutant patient fibroblasts, which showed impaired cell cycle arrest under HS and compromised survival at recovery. Taken together, our results suggest that the altered phosphorylation stoichiometry of lamin A/C provides an evolutionarily conserved mechanism to regulate lamina structure and serve nuclear adaptation and cell survival during HS.
Keywords: Heat shock; Heat shock response; Lamin A/C; Lap2α; Phosphorylation.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


References
-
- Buxboim, A., Swift, J., Irianto, J., Spinler, K. R., Dingal, P. C. D. P., Athirasala, A., Kao, Y. R. C., Cho, S., Harada, T., Shin, J. W.et al. (2014). Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr. Biol. 24, 1909-1917. 10.1016/j.cub.2014.07.001 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

