Data-Driven and Machine Learning-Based Framework for Image-Guided Single-Cell Mass Spectrometry
- PMID: 36695570
- PMCID: PMC9901547
- DOI: 10.1021/acs.jproteome.2c00714
Data-Driven and Machine Learning-Based Framework for Image-Guided Single-Cell Mass Spectrometry
Abstract
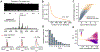
Improved throughput of analysis and lowered limits of detection have allowed single-cell chemical analysis to go beyond the detection of a few molecules in such volume-limited samples, enabling researchers to characterize different functional states of individual cells. Image-guided single-cell mass spectrometry leverages optical and fluorescence microscopy in the high-throughput analysis of cellular and subcellular targets. In this work, we propose DATSIGMA (DAta-driven Tools for Single-cell analysis using Image-Guided MAss spectrometry), a workflow based on data-driven and machine learning approaches for feature extraction and enhanced interpretability of complex single-cell mass spectrometry data. Here, we implemented our toolset with user-friendly programs and tested it on multiple experimental data sets that cover a wide range of biological applications, including classifying various brain cell types. Because it is open-source, it offers a high level of customization and can be easily adapted to other types of single-cell mass spectrometry data.
Keywords: data-driven analysis; machine learning; mass spectrometry; single-cell analysis.
Conflict of interest statement
CONFLICT OF INTEREST.
The authors declare no competing financial interest.
Figures





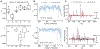

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

