Specific Plasma MicroRNA Signatures Underlying the Clinical Outcomes of Hepatitis E Virus Infection
- PMID: 36695578
- PMCID: PMC9927377
- DOI: 10.1128/spectrum.04664-22
Specific Plasma MicroRNA Signatures Underlying the Clinical Outcomes of Hepatitis E Virus Infection
Abstract
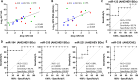
The pathogenic mechanisms determining the diverse clinical outcomes of HEV infection (e.g., self-limiting versus chronic or symptomatic versus asymptomatic) are not yet understood. Because specific microRNA signatures during viral infection inform the cellular processes involved in virus replication and pathogenesis, we investigated plasma microRNA profiles in 44 subjects, including patients with symptomatic acute (AHE, n = 7) and chronic (CHE, n = 6) hepatitis E, blood donors with asymptomatic infection (HEV BDs, n = 9), and anti-HEV IgG+ IgM- (exposed BDs, n = 10) and anti-HEV IgG- IgM- (naive BDs, n = 12) healthy blood donors. By measuring the abundance of 179 microRNAs in AHE patients and naive BDs by reverse transcription-quantitative PCR (RT-qPCR), we identified 51 potential HEV-regulated microRNAs (P value adjusted for multiple testing by the Benjamini-Hochberg correction [PBH] < 0.05). Further analysis showed that HEV genotype 3 infection is associated with miR-122, miR-194, miR-885, and miR-30a upregulation and miR-221, miR-223, and miR-27a downregulation. AHE patients showed significantly higher levels of miR-122 and miR-194 and lower levels of miR-221, miR-27a, and miR-335 than HEV BDs. This specific microRNA signature in AHE could promote virus replication and reduce antiviral immune responses, contributing to the development of clinical symptoms. We found that miR-194, miR-335, and miR-221 can discriminate between asymptomatic HEV infections and those developing acute symptoms, whereas miR-335 correctly classifies AHE and CHE patients. Our data suggest that diverse outcomes of HEV infection result from different HEV-induced microRNA dysregulations. The specific microRNA signatures described offer novel information that may serve to develop biomarkers of HEV infection outcomes and improve our understanding of HEV pathogenesis, which may facilitate the identification of antiviral targets. IMPORTANCE There is increasing evidence that viruses dysregulate the expression and/or secretion of microRNAs to promote viral replication, immune evasion, and pathogenesis. In this study, we evaluated the change in microRNA abundance in patients with acute or chronic HEV infection and asymptomatic HEV-infected blood donors. Our results suggest that different HEV-induced microRNA dysregulations may contribute to the diverse clinical manifestations of HEV infection. The specific microRNA signatures identified in this study hold potential as predictive markers of HEV infection outcomes, which would improve the clinical management of hepatitis E patients, particularly of those developing severe symptoms or chronic infections. Furthermore, this study provides new insights into HEV pathogenesis that may serve to identify antiviral targets, which would have a major impact because no effective treatments are yet available.
Keywords: diagnostics; hepatitis E virus; infection outcomes; microRNA; prognostic indicators.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Positive Regulation of Hepatitis E Virus Replication by MicroRNA-122.J Virol. 2018 May 14;92(11):e01999-17. doi: 10.1128/JVI.01999-17. Print 2018 Jun 1. J Virol. 2018. PMID: 29540601 Free PMC article.
-
Specific circulating microRNAs during hepatitis E infection can serve as indicator for chronic hepatitis E.Sci Rep. 2020 Mar 24;10(1):5337. doi: 10.1038/s41598-020-62159-9. Sci Rep. 2020. PMID: 32210284 Free PMC article.
-
Expression Profiles of Exosomal MicroRNAs from HEV- and HCV-Infected Blood Donors and Patients: A Pilot Study.Viruses. 2020 Jul 30;12(8):833. doi: 10.3390/v12080833. Viruses. 2020. PMID: 32751663 Free PMC article.
-
From discovery to treatment: tracing the path of hepatitis E virus.Virol J. 2024 Aug 23;21(1):194. doi: 10.1186/s12985-024-02470-3. Virol J. 2024. PMID: 39180020 Free PMC article. Review.
-
Hepatitis E virus in hematopoietic stem cell transplant recipients: A systematic review.J Clin Virol. 2019 Oct;119:31-36. doi: 10.1016/j.jcv.2019.08.002. Epub 2019 Aug 2. J Clin Virol. 2019. PMID: 31450185
Cited by
-
Differential miRNA signatures in Hepatitis E Virus Infection: Insights into acute, chronic, and pregnancy-related outcomes.Eur J Clin Microbiol Infect Dis. 2025 Jul 14. doi: 10.1007/s10096-025-05207-4. Online ahead of print. Eur J Clin Microbiol Infect Dis. 2025. PMID: 40660006 Review.
-
Role of microRNAs in chronic hepatitis E viral infection.Bioinformation. 2025 Feb 28;21(2):240-252. doi: 10.6026/973206300210240. eCollection 2025. Bioinformation. 2025. PMID: 40322700 Free PMC article.
-
Pathogenesis and clinical features of severe hepatitis E virus infection.World J Virol. 2024 Jun 25;13(2):91580. doi: 10.5501/wjv.v13.i2.91580. World J Virol. 2024. PMID: 38984076 Free PMC article. Review.
-
Hepatitis E Virus Infection Caused Elevation of Alanine Aminotransferase Levels in a Patient with Chronic Hepatitis B and Choledocholithiasis.Reports (MDPI). 2023 Nov 17;6(4):55. doi: 10.3390/reports6040055. Reports (MDPI). 2023. PMID: 40729178 Free PMC article.
References
-
- Smith DB, Simmonds P, Members Of The International Committee On The Taxonomy Of Viruses Study Group, Jameel S, Emerson SU, Harrison TJ, Meng X-J, Okamoto H, Van der Poel WHM, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95:2223–2232. doi:10.1099/vir.0.068429-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

