Genome-wide screen in human plasma identifies multifaceted complement evasion of Pseudomonas aeruginosa
- PMID: 36696456
- PMCID: PMC9901815
- DOI: 10.1371/journal.ppat.1011023
Genome-wide screen in human plasma identifies multifaceted complement evasion of Pseudomonas aeruginosa
Abstract
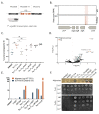
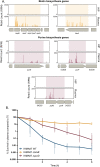
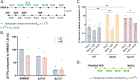
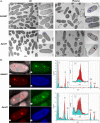
Pseudomonas aeruginosa, an opportunistic Gram-negative pathogen, is a leading cause of bacteremia with a high mortality rate. We recently reported that P. aeruginosa forms a persister-like sub-population of evaders in human plasma. Here, using a gain-of-function transposon sequencing (Tn-seq) screen in plasma, we identified and validated previously unknown factors affecting bacterial persistence in plasma. Among them, we identified a small periplasmic protein, named SrgA, whose expression leads to up to a 100-fold increase in resistance to killing. Additionally, mutants in pur and bio genes displayed higher tolerance and persistence, respectively. Analysis of several steps of the complement cascade and exposure to an outer-membrane-impermeable drug, nisin, suggested that the mutants impede membrane attack complex (MAC) activity per se. Electron microscopy combined with energy-dispersive X-ray spectroscopy (EDX) revealed the formation of polyphosphate (polyP) granules upon incubation in plasma of different size in purD and wild-type strains, implying the bacterial response to a stress signal. Indeed, inactivation of ppk genes encoding polyP-generating enzymes lead to significant elimination of persisting bacteria from plasma. Through this study, we shed light on a complex P. aeruginosa response to the plasma conditions and discovered the multifactorial origin of bacterial resilience to MAC-induced killing.
Copyright: © 2023 Janet-Maitre et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

