Minimal synthetic enhancers reveal control of the probability of transcriptional engagement and its timing by a morphogen gradient
- PMID: 36696901
- PMCID: PMC10125799
- DOI: 10.1016/j.cels.2022.12.008
Minimal synthetic enhancers reveal control of the probability of transcriptional engagement and its timing by a morphogen gradient
Abstract
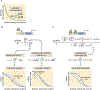
How enhancers interpret morphogen gradients to generate gene expression patterns is a central question in developmental biology. Recent studies have proposed that enhancers can dictate whether, when, and at what rate promoters engage in transcription, but the complexity of endogenous enhancers calls for theoretical models with too many free parameters to quantitatively dissect these regulatory strategies. To overcome this limitation, we established a minimal promoter-proximal synthetic enhancer in embryos of Drosophila melanogaster. Here, a gradient of the Dorsal activator is read by a single Dorsal DNA binding site. Using live imaging to quantify transcriptional activity, we found that a single binding site can regulate whether promoters engage in transcription in a concentration-dependent manner. By modulating the binding-site affinity, we determined that a gene's decision to transcribe and its transcriptional onset time can be explained by a simple model where the promoter traverses multiple kinetic barriers before transcription can ensue.
Keywords: Drosophila melanogaster; biophysics; developmental biology; quantitative biology; transcriptional dynamic; transcriptional modeling; transcriptional regulation.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
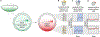





Similar articles
-
Predictive modeling reveals that higher-order cooperativity drives transcriptional repression in a synthetic developmental enhancer.Elife. 2022 Dec 12;11:e73395. doi: 10.7554/eLife.73395. Elife. 2022. PMID: 36503705 Free PMC article.
-
Regulation of spatiotemporal limits of developmental gene expression via enhancer grammar.Proc Natl Acad Sci U S A. 2020 Jun 30;117(26):15096-15103. doi: 10.1073/pnas.1917040117. Epub 2020 Jun 15. Proc Natl Acad Sci U S A. 2020. PMID: 32541043 Free PMC article.
-
Two promoters integrate multiple enhancer inputs to drive wild-type knirps expression in the Drosophila melanogaster embryo.Genetics. 2021 Dec 10;219(4):iyab154. doi: 10.1093/genetics/iyab154. Genetics. 2021. PMID: 34849867 Free PMC article.
-
One thousand and one ways of making functionally similar transcriptional enhancers.Bioessays. 2008 Nov;30(11-12):1052-7. doi: 10.1002/bies.20849. Bioessays. 2008. PMID: 18937349 Review.
-
Dynamic patterning by morphogens illuminated by cis-regulatory studies.Development. 2021 Jan 20;148(2):dev196113. doi: 10.1242/dev.196113. Development. 2021. PMID: 33472851 Free PMC article. Review.
Cited by
-
Deciphering the multi-scale, quantitative cis-regulatory code.Mol Cell. 2023 Feb 2;83(3):373-392. doi: 10.1016/j.molcel.2022.12.032. Epub 2023 Jan 23. Mol Cell. 2023. PMID: 36693380 Free PMC article. Review.
-
A bistable autoregulatory module in the developing embryo commits cells to binary expression fates.Curr Biol. 2023 Jul 24;33(14):2851-2864.e11. doi: 10.1016/j.cub.2023.06.060. Epub 2023 Jul 14. Curr Biol. 2023. PMID: 37453424 Free PMC article.
-
Novel Fluorescent and Photoconvertible Fusions Reveal Dorsal Activator Dynamics.bioRxiv [Preprint]. 2025 Jul 23:2025.05.12.653543. doi: 10.1101/2025.05.12.653543. bioRxiv. 2025. PMID: 40462965 Free PMC article. Preprint.
-
Competing constraints shape the nonequilibrium limits of cellular decision-making.Proc Natl Acad Sci U S A. 2023 Mar 7;120(10):e2211203120. doi: 10.1073/pnas.2211203120. Epub 2023 Mar 2. Proc Natl Acad Sci U S A. 2023. PMID: 36862689 Free PMC article.
-
Time will tell: comparing timescales to gain insight into transcriptional bursting.Trends Genet. 2024 Feb;40(2):160-174. doi: 10.1016/j.tig.2023.11.003. Epub 2024 Jan 12. Trends Genet. 2024. PMID: 38216391 Free PMC article. Review.
References
-
- Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol, 25(1):1–47, 1969. - PubMed
-
- Garcia HG, Berrocal A, Kim YJ, Martini G, and Zhao J. Lighting up the central dogma for predictive developmental biology. Curr Top Dev Biol, 137:1–35, 2020. - PubMed
-
- Vincent BJ, Estrada J, and DePace AH. The appeasement of doug: a synthetic approach to enhancer biology. Integr Biol (Camb), 8(4):475–84, 2016. - PubMed
-
- Venken Koen J. T. and Bellen Hugo J.. Emerging technologies for gene manipulation in Drosophila melanogaster. Nature Reviews Genetics, 6(3):167–178, March 2005. Number: 3 Publisher: Nature Publishing Group. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

