3'UTR Diversity: Expanding Repertoire of RNA Alterations in Human mRNAs
- PMID: 36697237
- PMCID: PMC9880603
- DOI: 10.14348/molcells.2023.0003
3'UTR Diversity: Expanding Repertoire of RNA Alterations in Human mRNAs
Abstract
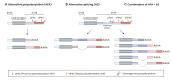
Genomic information stored in the DNA is transcribed to the mRNA and translated to proteins. The 3' untranslated regions (3'UTRs) of the mRNA serve pivotal roles in posttranscriptional gene expression, regulating mRNA stability, translation, and localization. Similar to DNA mutations producing aberrant proteins, RNA alterations expand the transcriptome landscape and change the cellular proteome. Recent global analyses reveal that many genes express various forms of altered RNAs, including 3'UTR length variants. Alternative polyadenylation and alternative splicing are involved in diversifying 3'UTRs, which could act as a hidden layer of eukaryotic gene expression control. In this review, we summarize the functions and regulations of 3'UTRs and elaborate on the generation and functional consequences of 3'UTR diversity. Given that dynamic 3'UTR length control contributes to phenotypic complexity, dysregulated 3'UTR diversity might be relevant to disease development, including cancers. Thus, 3'UTR diversity in cancer could open exciting new research areas and provide avenues for novel cancer theragnostics.
Keywords: 3′UTR diversity; RNA alterations; alternative polyadenylation; alternative splicing; cancer; transcriptome.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

