Artificial intelligence applied in neoantigen identification facilitates personalized cancer immunotherapy
- PMID: 36698417
- PMCID: PMC9868469
- DOI: 10.3389/fonc.2022.1054231
Artificial intelligence applied in neoantigen identification facilitates personalized cancer immunotherapy
Abstract
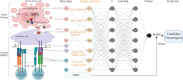
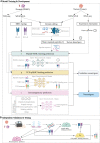
The field of cancer neoantigen investigation has developed swiftly in the past decade. Predicting novel and true neoantigens derived from large multi-omics data became difficult but critical challenges. The rise of Artificial Intelligence (AI) or Machine Learning (ML) in biomedicine application has brought benefits to strengthen the current computational pipeline for neoantigen prediction. ML algorithms offer powerful tools to recognize the multidimensional nature of the omics data and therefore extract the key neoantigen features enabling a successful discovery of new neoantigens. The present review aims to outline the significant technology progress of machine learning approaches, especially the newly deep learning tools and pipelines, that were recently applied in neoantigen prediction. In this review article, we summarize the current state-of-the-art tools developed to predict neoantigens. The standard workflow includes calling genetic variants in paired tumor and blood samples, and rating the binding affinity between mutated peptide, MHC (I and II) and T cell receptor (TCR), followed by characterizing the immunogenicity of tumor epitopes. More specifically, we highlight the outstanding feature extraction tools and multi-layer neural network architectures in typical ML models. It is noted that more integrated neoantigen-predicting pipelines are constructed with hybrid or combined ML algorithms instead of conventional machine learning models. In addition, the trends and challenges in further optimizing and integrating the existing pipelines are discussed.
Keywords: artificial intelligence; cancer immunotherapy; cancer neoantigen; neoantigen prediction; next generation sequencing.
Copyright © 2023 Cai, Chen, Gao, Li, Liu, Su, Song, Jiang, Jiang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Bhinder B, Gilvary C, Madhukar NS, Elemento O. Artificial intelligence in cancer research and precision medicine. Cancer Discovery (2021) 11(4):900–15. doi: 10.1158/2159-8290.Cd-21-0090 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

