Prediction of antibody binding to SARS-CoV-2 RBDs
- PMID: 36698760
- PMCID: PMC9868522
- DOI: 10.1093/bioadv/vbac103
Prediction of antibody binding to SARS-CoV-2 RBDs
Abstract
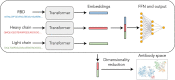
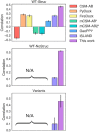
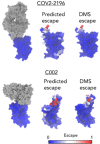
Summary: The ability to predict antibody-antigen binding is essential for computational models of antibody affinity maturation and protein design. While most models aim to predict binding for arbitrary antigens and antibodies, the global impact of SARS-CoV-2 on public health and the availability of associated data suggest that a SARS-CoV-2-specific model would be highly beneficial. In this work, we present a neural network model, trained on ∼315 000 datapoints from deep mutational scanning experiments, that predicts escape fractions of SARS-CoV-2 RBDs binding to arbitrary antibodies. The antibody embeddings within the model constitute an effective sequence space, which correlates with the Hamming distance, suggesting that these embeddings may be useful for downstream tasks such as binding prediction. Indeed, the model achieves Spearman correlation coefficients of 0.46 and 0.52 on two held-out test sets. By comparison, correlation coefficients calculated using existing structure and sequence-based models do not exceed 0.28. The correlation coefficient against dissociation constants of antibodies binding to SARS-CoV-2 RBD variants is 0.46. Additionally, the residue-level escapes are highest in the antibody epitope, correlating well with experimentally measured escapes. We further study the effect of antibody chain use, embedding dimension size and feed-forward and convolutional architectures on the model results. Lastly, we find that the inference time of our model is significantly faster than previous models, suggesting that it could be a useful tool for the accurate and rapid prediction of antibodies binding to SARS-CoV-2 RBDs.
Availability and implementation: The model and associated code are available for download at https://github.com/ericzwang/RBD_AB.
Supplementary information: Supplementary data are available at Bioinformatics Advances online.
© The Author(s) 2023. Published by Oxford University Press.
Figures



Similar articles
-
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection.Elife. 2022 Jan 24;11:e73490. doi: 10.7554/eLife.73490. Elife. 2022. PMID: 35072628 Free PMC article.
-
Computational Analysis of Mutations in the Receptor-Binding Domain of SARS-CoV-2 Spike and Their Effects on Antibody Binding.Viruses. 2022 Jan 30;14(2):295. doi: 10.3390/v14020295. Viruses. 2022. PMID: 35215888 Free PMC article.
-
Mechanistic Insights to the Binding of Antibody CR3022 Against RBD from SARS-CoV and HCoV-19/SARS-CoV-2: A Computational Study.Comb Chem High Throughput Screen. 2021;24(7):1069-1082. doi: 10.2174/1386207323666201026160500. Comb Chem High Throughput Screen. 2021. PMID: 33106140
-
MVSF-AB: accurate antibody-antigen binding affinity prediction via multi-view sequence feature learning.Bioinformatics. 2025 May 6;41(5):btae579. doi: 10.1093/bioinformatics/btae579. Bioinformatics. 2025. PMID: 39363630 Free PMC article.
-
SARS-CoV-2: Recent Variants and Clinical Efficacy of Antibody-Based Therapy.Front Cell Infect Microbiol. 2022 Feb 14;12:839170. doi: 10.3389/fcimb.2022.839170. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35237535 Free PMC article. Review.
Cited by
-
NABP-BERT: NANOBODY®-antigen binding prediction based on bidirectional encoder representations from transformers (BERT) architecture.Brief Bioinform. 2024 Nov 22;26(1):bbae518. doi: 10.1093/bib/bbae518. Brief Bioinform. 2024. PMID: 39688476 Free PMC article.
-
In silico design of immunogenic antigen cocktail via affinity maturation-guided optimization.Bioinform Adv. 2025 Jul 28;5(1):vbaf182. doi: 10.1093/bioadv/vbaf182. eCollection 2025. Bioinform Adv. 2025. PMID: 40831759 Free PMC article.
-
Generative prediction of real-world prevalent SARS-CoV-2 mutation with in silico virus evolution.Brief Bioinform. 2025 May 1;26(3):bbaf276. doi: 10.1093/bib/bbaf276. Brief Bioinform. 2025. PMID: 40532108 Free PMC article.
-
Exploring the ability of the MD+FoldX method to predict SARS-CoV-2 antibody escape mutations using large-scale data.bioRxiv [Preprint]. 2024 May 22:2024.05.22.595230. doi: 10.1101/2024.05.22.595230. bioRxiv. 2024. Update in: Sci Rep. 2024 Oct 4;14(1):23122. doi: 10.1038/s41598-024-72491-z. PMID: 38826284 Free PMC article. Updated. Preprint.
-
Biophysics of SARS-CoV-2 spike protein's receptor-binding domain interaction with ACE2 and neutralizing antibodies: from computation to functional insights.Biophys Rev. 2025 Mar 8;17(2):309-333. doi: 10.1007/s12551-025-01276-z. eCollection 2025 Apr. Biophys Rev. 2025. PMID: 40376405 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

