Acute and chronic HBV infection in central Argentina: High frequency of sub-genotype F1b, low detection of clinically relevant mutations and first evidence of HDV
- PMID: 36698842
- PMCID: PMC9868314
- DOI: 10.3389/fmed.2022.1057194
Acute and chronic HBV infection in central Argentina: High frequency of sub-genotype F1b, low detection of clinically relevant mutations and first evidence of HDV
Abstract
Introduction: Genomic analysis of hepatitis B virus (HBV) identifies phylogenetic variants, which may lead to distinct biological and clinical behaviors. The satellite hepatitis D virus (HDV) may also influence clinical outcomes in patients with hepatitis B. The aim of this study was to investigate HBV genetic variants, including clinically relevant mutations, and HDV infection in acute and chronic hepatitis B patients in central Argentina.
Methods: A total of 217 adult HBV infected patients [acute (AHB): n = 79; chronic (CHB): n = 138] were studied; 67 were HBV/human immunodeficiency virus (HIV) coinfected. Clinical and demographic data were obtained from medical records. Serological markers were determined. Molecular detection of HBV and HDV was carried out by RT-Nested PCR, followed by sequencing and phylogenetic analysis.
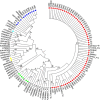
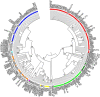
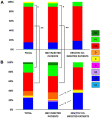
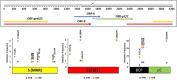
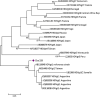
Results: Overall, genotype (gt) F [sub-genotype (sgt) F1b] was the most frequently found. In AHB patients, the gts/sgts found were: F1b (74.7%) > A2 (13.9%) > F4 (7.6%) > C (2.5%) > A1 (1.3%). Among CHB patients: F1b (39.1%) > A2 (23.9%) > F4 (18.2%) > D (9.4%) > C and F6 (3.6% each) > A1, A3 and B2 (0.7% each). The distribution of sgt A2 and gt D was significantly different between HBV mono and HBV/HIV coinfected patients [A2: 15.9% vs. 35.7% (p < 0.05), respectively and D: 14.6% vs. 1.8% (p < 0.05), respectively]. Mutation frequency in basal core promoter/pre-Core (BCP/pC) region was 35.5% (77/217) [AHB: 20.3% (16/79), CHB: 44.2% (61/138)]. In the open reading frame (ORF) S, mutations associated with vaccine escape and diagnostic failure were detected in 7.8% of the sequences (17/217) [AHB: 3.8% (3/79), CHB: 10.1% (14/138)]. ORF-P amino acid substitutions associated with antiviral resistance were detected in 3.2% of the samples (7/217) [AHB: 1.3% (1/79), CHB 4.3%, (6/138)]. The anti-HDV seropositivity was 5.2% (4/77); one sample could be sequenced, belonging to gt HDV-1 associated with sgt HBV-D3.
Discussion: We detected an increase in the circulation of genotype F in Central Argentina, particularly among AHB patients, suggesting transmission advantages over the other genotypes. A low rate of mutations was detected, especially those with antiviral resistance implications, which is an encouraging result. The evidence of HDV circulation in our region, reported for the first time, alerts the health system for its search and diagnosis.
Keywords: Argentina; HBV; antiviral resistance; genotypes; hepatitis B virus; mutant.
Copyright © 2023 Castro, Sosa, Sicilia, Riberi, Moreno, Cattaneo, Debes, Barbás, Cudolá, Pisano and Ré.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Features and clinical implications of hepatitis B virus genotypes and mutations in basal core promoter/precore region in 507 Chinese patients with acute and chronic hepatitis B.J Clin Virol. 2010 Mar;47(3):243-7. doi: 10.1016/j.jcv.2009.12.013. Epub 2010 Jan 15. J Clin Virol. 2010. PMID: 20080060
-
Molecular epidemiology of hepatitis B virus in Córdoba, Argentina.J Clin Virol. 2014 Oct;61(2):204-10. doi: 10.1016/j.jcv.2014.06.030. Epub 2014 Jul 9. J Clin Virol. 2014. PMID: 25066884
-
Virological Characteristics of Acute Hepatitis B in Eastern India: Critical Differences with Chronic Infection.PLoS One. 2015 Nov 16;10(11):e0141741. doi: 10.1371/journal.pone.0141741. eCollection 2015. PLoS One. 2015. PMID: 26571502 Free PMC article.
-
Drug-resistant and immune-escape hepatitis B virus mutants, occult hepatitis B infection and coinfections in public hospital patients from Argentina.Virus Genes. 2021 Aug;57(4):327-337. doi: 10.1007/s11262-021-01850-z. Epub 2021 Jun 6. Virus Genes. 2021. PMID: 34091827
-
Molecular epidemiology and genetic diversity of hepatitis B virus in Mar del Plata city, Argentina.Infect Genet Evol. 2013 Oct;19:152-63. doi: 10.1016/j.meegid.2013.07.007. Epub 2013 Jul 17. Infect Genet Evol. 2013. PMID: 23871776
References
-
- World Health Organization [WHO]. Home/Newsroom/Fact sheets/Detail/Hepatitis B. Geneva: WHO; (2022).
-
- Pujol F, Jaspe R, Loureiro C, Chemin I. B virus American genotypes: pathogenic variants? Clin Res Hepatol Gastroenterol. (2020) 44:825–35. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

