Genetic diversity in global populations of the critically endangered addax (Addax nasomaculatus) and its implications for conservation
- PMID: 36699120
- PMCID: PMC9850015
- DOI: 10.1111/eva.13515
Genetic diversity in global populations of the critically endangered addax (Addax nasomaculatus) and its implications for conservation
Abstract
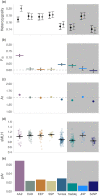
Threatened species are frequently patchily distributed across small wild populations, ex situ populations managed with varying levels of intensity and reintroduced populations. Best practice advocates for integrated management across in situ and ex situ populations. Wild addax (Addax nasomaculatus) now number fewer than 100 individuals, yet 1000 of addax remain in ex situ populations, which can provide addax for reintroductions, as has been the case in Tunisia since the mid-1980s. However, integrated management requires genetic data to ascertain the relationships between wild and ex situ populations that have incomplete knowledge of founder origins, management histories, and pedigrees. We undertook a global assessment of genetic diversity across wild, ex situ and reintroduced populations in Tunisia to assist conservation planning for this Critically Endangered species. We show that the remnant wild populations retain more mitochondrial haplotypes that are more diverse than the entirety of the ex situ populations across Europe, North America and the United Arab Emirates, and the reintroduced Tunisian population. Additionally, 1704 SNPs revealed that whilst population structure within the ex situ population is minimal, each population carries unique diversity. Finally, we show that careful selection of founders and subsequent genetic management is vital to ensure genetic diversity is provided to, and minimize drift and inbreeding within reintroductions. Our results highlight a vital need to conserve the last remaining wild addax population, and we provide a genetic foundation for determining integrated conservation strategies to prevent extinction and optimize future reintroductions.
Keywords: Addax nasomaculatus; Sahelo‐Saharan antelope; captive populations; conservation genetics; reintroduction; ungulate conservation.
© 2022 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests in relation to this manuscript.
Figures



Similar articles
-
Diversity and Paleodemography of the Addax (Addax nasomaculatus), a Saharan Antelope on the Verge of Extinction.Genes (Basel). 2021 Aug 11;12(8):1236. doi: 10.3390/genes12081236. Genes (Basel). 2021. PMID: 34440410 Free PMC article.
-
A suite of microsatellite markers optimized for amplification of DNA from Addax (Addax nasomaculatus) blood preserved on FTA cards.Zoo Biol. 2012 Jan-Feb;31(1):98-106. doi: 10.1002/zoo.20420. Epub 2011 Sep 6. Zoo Biol. 2012. PMID: 21898577
-
Reprint of "Echinococcus granulosus sensu stricto (s.s.) from the critically endangered antelope Addax nasomaculatus in Tunisia".Acta Trop. 2017 Jan;165:17-20. doi: 10.1016/j.actatropica.2015.08.021. Epub 2016 Nov 23. Acta Trop. 2017. PMID: 27887695
-
Meta-analysis of genetic representativeness of plant populations under ex situ conservation in contrast to wild source populations.Conserv Biol. 2020 Aug 24. doi: 10.1111/cobi.13617. Online ahead of print. Conserv Biol. 2020. PMID: 32840007 Review.
-
Quantitative genetics in conservation biology.Genet Res. 1999 Dec;74(3):237-44. doi: 10.1017/s001667239900405x. Genet Res. 1999. PMID: 10689801 Review.
Cited by
-
The environment matters: season and female contact affect the behavior of captive Addax nasomaculatus male antelope.Acta Ethol. 2023;26(2):109-117. doi: 10.1007/s10211-023-00419-3. Epub 2023 Apr 6. Acta Ethol. 2023. PMID: 37261311 Free PMC article.
-
Genomics identifies koala populations at risk across eastern Australia.Ecol Appl. 2025 Jan;35(1):e3062. doi: 10.1002/eap.3062. Epub 2024 Nov 29. Ecol Appl. 2025. PMID: 39611546 Free PMC article.
-
The genetic legacy of the first successful reintroduction of a mammal to Britain: Founder events and attempted genetic rescue in Scotland's beaver population.Evol Appl. 2023 Dec 28;17(2):e13629. doi: 10.1111/eva.13629. eCollection 2024 Feb. Evol Appl. 2023. PMID: 38343777 Free PMC article.
-
Genome-wide diversity evaluation and core germplasm extraction in ex situ conservation: A case of golden Camellia tunghinensis.Evol Appl. 2023 Aug 19;16(9):1519-1530. doi: 10.1111/eva.13584. eCollection 2023 Sep. Evol Appl. 2023. PMID: 37752963 Free PMC article.
References
-
- Aktas, C. (2020). haplotypes: Manipulating DNA sequences and estimating unambiguous haplotype network with statistical parsimony . (R package version 1.1.2). https://cran.r‐project.org/package=haplotypes
-
- Allendorf, F. W. (1986). Genetic drift and the loss of alleles versus heterozygosity. Zoo Biology, 5, 181–190. 10.1002/ZOO.1430050212 - DOI
-
- Allendorf, F. W. , & Luikart, G. (2006). Conservation and the genetics of populations. Blackwell Publishing.
-
- Armstrong, E. , Leizagoyen, C. , Martínez, A. M. , González, S. , Delgado, J. V. , & Postiglioni, A. (2011). Genetic structure analysis of a highly inbred captive population of the African antelope Addax nasomaculatus. Conservation and management implications. Zoo Biology, 30, 399–411. 10.1002/zoo.20341 - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources

