Genetic diversity in global populations of the critically endangered addax (Addax nasomaculatus) and its implications for conservation
- PMID: 36699120
- PMCID: PMC9850015
- DOI: 10.1111/eva.13515
Genetic diversity in global populations of the critically endangered addax (Addax nasomaculatus) and its implications for conservation
Abstract
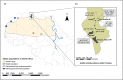
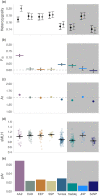
Threatened species are frequently patchily distributed across small wild populations, ex situ populations managed with varying levels of intensity and reintroduced populations. Best practice advocates for integrated management across in situ and ex situ populations. Wild addax (Addax nasomaculatus) now number fewer than 100 individuals, yet 1000 of addax remain in ex situ populations, which can provide addax for reintroductions, as has been the case in Tunisia since the mid-1980s. However, integrated management requires genetic data to ascertain the relationships between wild and ex situ populations that have incomplete knowledge of founder origins, management histories, and pedigrees. We undertook a global assessment of genetic diversity across wild, ex situ and reintroduced populations in Tunisia to assist conservation planning for this Critically Endangered species. We show that the remnant wild populations retain more mitochondrial haplotypes that are more diverse than the entirety of the ex situ populations across Europe, North America and the United Arab Emirates, and the reintroduced Tunisian population. Additionally, 1704 SNPs revealed that whilst population structure within the ex situ population is minimal, each population carries unique diversity. Finally, we show that careful selection of founders and subsequent genetic management is vital to ensure genetic diversity is provided to, and minimize drift and inbreeding within reintroductions. Our results highlight a vital need to conserve the last remaining wild addax population, and we provide a genetic foundation for determining integrated conservation strategies to prevent extinction and optimize future reintroductions.
Keywords: Addax nasomaculatus; Sahelo‐Saharan antelope; captive populations; conservation genetics; reintroduction; ungulate conservation.
© 2022 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests in relation to this manuscript.
Figures



References
-
- Aktas, C. (2020). haplotypes: Manipulating DNA sequences and estimating unambiguous haplotype network with statistical parsimony . (R package version 1.1.2). https://cran.r‐project.org/package=haplotypes
-
- Allendorf, F. W. (1986). Genetic drift and the loss of alleles versus heterozygosity. Zoo Biology, 5, 181–190. 10.1002/ZOO.1430050212 - DOI
-
- Allendorf, F. W. , & Luikart, G. (2006). Conservation and the genetics of populations. Blackwell Publishing.
-
- Armstrong, E. , Leizagoyen, C. , Martínez, A. M. , González, S. , Delgado, J. V. , & Postiglioni, A. (2011). Genetic structure analysis of a highly inbred captive population of the African antelope Addax nasomaculatus. Conservation and management implications. Zoo Biology, 30, 399–411. 10.1002/zoo.20341 - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources

