Translational control in cortical development
- PMID: 36699134
- PMCID: PMC9868627
- DOI: 10.3389/fnana.2022.1087949
Translational control in cortical development
Abstract
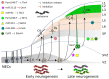
Differentiation of specific neuronal types in the nervous system is worked out through a complex series of gene regulation events. Within the mammalian neocortex, the appropriate expression of key transcription factors allocates neurons to different cortical layers according to an inside-out model and endows them with specific properties. Precise timing is required to ensure the proper sequential appearance of key transcription factors that dictate the identity of neurons within the different cortical layers. Recent evidence suggests that aspects of this time-controlled regulation of gene products rely on post-transcriptional control, and point at micro-RNAs (miRs) and RNA-binding proteins as important players in cortical development. Being able to simultaneously target many different mRNAs, these players may be involved in controlling the global expression of gene products in progenitors and post-mitotic cells, in a gene expression framework where parallel to transcriptional gene regulation, a further level of control is provided to refine and coordinate the appearance of the final protein products. miRs and RNA-binding proteins (RBPs), by delaying protein appearance, may play heterochronic effects that have recently been shown to be relevant for the full differentiation of cortical neurons and for their projection abilities. Such heterochronies may be the base for evolutionary novelties that have enriched the spectrum of cortical cell types within the mammalian clade.
Keywords: RNA binding protein; cortex; development; evolution; microRNA.
Copyright © 2023 Cremisi and Vignali.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Evolution of the Neocortex Through RNA-Binding Proteins and Post-transcriptional Regulation.Front Neurosci. 2022 Jan 10;15:803107. doi: 10.3389/fnins.2021.803107. eCollection 2021. Front Neurosci. 2022. PMID: 35082597 Free PMC article. Review.
-
RNA on the brain: emerging layers of post-transcriptional regulation in cerebral cortex development.Wiley Interdiscip Rev Dev Biol. 2018 Jan;7(1):10.1002/wdev.290. doi: 10.1002/wdev.290. Epub 2017 Aug 24. Wiley Interdiscip Rev Dev Biol. 2018. PMID: 28837264 Free PMC article. Review.
-
Post-transcriptional regulation in corticogenesis: how RNA-binding proteins help build the brain.Wiley Interdiscip Rev RNA. 2015 Sep-Oct;6(5):501-15. doi: 10.1002/wrna.1289. Epub 2015 Jun 18. Wiley Interdiscip Rev RNA. 2015. PMID: 26088328 Free PMC article. Review.
-
Cited2 Regulates Neocortical Layer II/III Generation and Somatosensory Callosal Projection Neuron Development and Connectivity.J Neurosci. 2016 Jun 15;36(24):6403-19. doi: 10.1523/JNEUROSCI.4067-15.2016. J Neurosci. 2016. PMID: 27307230 Free PMC article.
-
Pum2 and TDP-43 refine area-specific cytoarchitecture post-mitotically and modulate translation of Sox5, Bcl11b, and Rorb mRNAs in developing mouse neocortex.Elife. 2022 Mar 9;11:e55199. doi: 10.7554/eLife.55199. Elife. 2022. PMID: 35262486 Free PMC article.
References
LinkOut - more resources
Full Text Sources

