Circ-ATIC regulates esophageal squamous cell carcinoma growth and metastasis through miR-1294/PBX3 pathway
- PMID: 36699282
- PMCID: PMC9868444
- DOI: 10.1016/j.heliyon.2023.e12916
Circ-ATIC regulates esophageal squamous cell carcinoma growth and metastasis through miR-1294/PBX3 pathway
Abstract
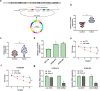
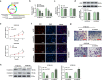
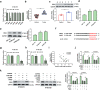
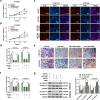
Esophageal squamous cell carcinoma (ESCC) is a digestive tract malignancy associated with poor clinical outcome. Growing evidence have elucidated that circular RNAs (circRNAs) play important roles in the pathological process of ESCC. However, the detailed mechanisms how circRNAs modulate the development of ESCC remain largely unknown. Our study aimed to decipher the role and mechanism of circ-ATIC (also termed as circRNA_0058063) in regulating the progression of ESCC. We found that circ-ATIC and its host gene ATIC were significantly increased in ESCC tissues and cells compared with the adjacent noncancerous tissues or normal esophagus epithelial cell. Circ-ATIC knockdown substantially reduced proliferation and the number of invaded ESCC cells and retarded EMT process, reflecting by the decreased N-cadherin and elevated E-cadherin. However, the level of host gene ATIC was not changed under circ-ATIC suppression. It was predicted that circ-ATIC could bind to miR-1294 and serve as a sponge RNA. The luciferase reporter assay and RNA immunoprecipitation (RIP) assay confirmed their relations. MiR-1294 was decreased in ESCC tissues and cells, which was reversely correlated with circ-ATIC level. Furthermore, PBX3 was predicted and proved to be a downstream direct target of miR-1294. PBX3 mRNA and protein were obviously upregulated in ESCC tumor tissues and cells. PBX3 overexpression could reverse the suppressive roles of miR-1294 mimics on ESCC proliferation and invasion. In an xenograft nude mice model, stable transfection of sh-circ-ATIC significantly retarded the growth of tumor and suppressed VEGF and Ki67. Collectively, circ-ATIC promoted ESCC proliferation and invasion by regulating miR-1294/PBX3 axis.
Keywords: Circ-ATIC; Esophageal squamous cell carcinoma; MiR-1294; PBX3.
© 2023 The Authors.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



Similar articles
-
Circ-ATIC Serves as a Sponge of miR-326 to Accelerate Esophageal Squamous Cell Carcinoma Progression by Targeting ID1.Biochem Genet. 2022 Oct;60(5):1585-1600. doi: 10.1007/s10528-021-10167-3. Epub 2022 Jan 22. Biochem Genet. 2022. PMID: 35064360
-
miR-542-3p/PIK3R1 axis is involved in hsa_circ_0087104-mediated inhibition of esophageal squamous cell carcinoma metastasis.Am J Cancer Res. 2024 Dec 15;14(12):5665-5679. doi: 10.62347/EFEO7205. eCollection 2024. Am J Cancer Res. 2024. PMID: 39803650 Free PMC article.
-
Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway.IUBMB Life. 2020 Mar;72(3):426-439. doi: 10.1002/iub.2202. Epub 2019 Nov 28. IUBMB Life. 2020. PMID: 31778020
-
Hsa_circ_0023984 Regulates Cell Proliferation, Migration, and Invasion in Esophageal Squamous Cancer via Regulating miR-1294/PI3K/Akt/c-Myc Pathway.Appl Biochem Biotechnol. 2022 Sep;194(9):1-16. doi: 10.1007/s12010-022-03935-3. Epub 2022 May 6. Appl Biochem Biotechnol. 2022. PMID: 35522362
-
Current advances and future perspectives on the functional roles and clinical implications of circular RNAs in esophageal squamous cell carcinoma: more influential than expected.Biomark Res. 2022 Jun 7;10(1):41. doi: 10.1186/s40364-022-00388-y. Biomark Res. 2022. PMID: 35672804 Free PMC article. Review.
Cited by
-
Circ_0096710 facilitates tumor growth via controlling ADAM10 expression in esophageal squamous cell carcinoma.Thorac Cancer. 2025 Jan;16(1):e15483. doi: 10.1111/1759-7714.15483. Epub 2024 Dec 2. Thorac Cancer. 2025. PMID: 39620490 Free PMC article.
-
circRNA6448-14/miR-455-3p/OTUB2 axis stimulates glycolysis and stemness of esophageal squamous cell carcinoma.Aging (Albany NY). 2024 May 30;16(11):9485-9497. doi: 10.18632/aging.205879. Epub 2024 May 30. Aging (Albany NY). 2024. PMID: 38819228 Free PMC article.
-
PHAX enhanced LIN28B-mediated PBX3 mRNA stability to promote esophageal cancer development.Cancer Sci. 2025 Mar;116(3):808-823. doi: 10.1111/cas.16420. Epub 2024 Dec 12. Cancer Sci. 2025. PMID: 39668567 Free PMC article.
-
Uncovering the role of microRNAs in esophageal cancer: from pathogenesis to clinical applications.Front Pharmacol. 2025 Jan 29;16:1532558. doi: 10.3389/fphar.2025.1532558. eCollection 2025. Front Pharmacol. 2025. PMID: 39944625 Free PMC article. Review.
-
Roles of non-coding RNAs in the metabolism and pathogenesis of bladder cancer.Hum Cell. 2023 Jul;36(4):1343-1372. doi: 10.1007/s13577-023-00915-5. Epub 2023 May 20. Hum Cell. 2023. PMID: 37209205 Review.
References
-
- Matsuda S., Kawakubo H., Okamura A., Takahashi K., Toihata T., Takemura R., Mayanagi S., Hirata K., Irino T., Hamamoto Y., Takeuchi H., Watanabe M., Kitagawa Y. Distribution of residual disease and recurrence patterns in pathological responders after neoadjuvant chemotherapy for esophageal squamous cell carcinoma. Ann. Surg. 2020 doi: 10.1097/SLA.0000000000004436. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

