scMoC: single-cell multi-omics clustering
- PMID: 36699396
- PMCID: PMC9710707
- DOI: 10.1093/bioadv/vbac011
scMoC: single-cell multi-omics clustering
Abstract
Motivation: Single-cell multi-omics assays simultaneously measure different molecular features from the same cell. A key question is how to benefit from the complementary data available and perform cross-modal clustering of cells.
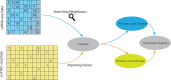
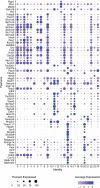
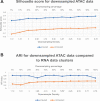
Results: We propose Single-Cell Multi-omics Clustering (scMoC), an approach to identify cell clusters from data with comeasurements of scRNA-seq and scATAC-seq from the same cell. We overcome the high sparsity of the scATAC-seq data by using an imputation strategy that exploits the less-sparse scRNA-seq data available from the same cell. Subsequently, scMoC identifies clusters of cells by merging clusterings derived from both data domains individually. We tested scMoC on datasets generated using different protocols with variable data sparsity levels. We show that scMoC (i) is able to generate informative scATAC-seq data due to its RNA-guided imputation strategy and (ii) results in integrated clusters based on both RNA and ATAC information that are biologically meaningful either from the RNA or from the ATAC perspective.
Availability and implementation: The data used in this manuscript is publicly available, and we refer to the original manuscript for their description and availability. For convience sci-CAR data is available at NCBI GEO under the accession number of GSE117089. SNARE-seq data is available at NCBI GEO under the accession number of GSE126074. The 10X multiome data is available at the following link https://www.10xgenomics.com/resources/datasets/pbmc-from-a-healthy-donor-no-cell-sorting-3-k-1-standard-2-0-0.
Supplementary information: Supplementary data are available at Bioinformatics Advances online.
© The Author(s) 2022. Published by Oxford University Press.
Figures



References
-
- Blondel V.D. et al. (2008) Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp., 2008, P10008.
LinkOut - more resources
Full Text Sources

