Museum specimens of a landlocked pinniped reveal recent loss of genetic diversity and unexpected population connections
- PMID: 36699566
- PMCID: PMC9849707
- DOI: 10.1002/ece3.9720
Museum specimens of a landlocked pinniped reveal recent loss of genetic diversity and unexpected population connections
Abstract
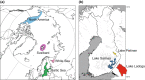
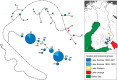
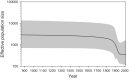
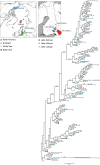
The Saimaa ringed seal (Pusa hispida saimensis) is endemic to Lake Saimaa in Finland. The subspecies is thought to have originated when parts of the ringed seal population of the Baltic region were trapped in lakes emerging due to postglacial bedrock rebound around 9000 years ago. During the 20th century, the population experienced a drastic human-induced bottleneck. Today encompassing a little over 400 seals with extremely low genetic diversity, it is classified as endangered. We sequenced sections of the mitochondrial control region from 60 up to 125-years-old museum specimens of the Saimaa ringed seal. The generated dataset was combined with publicly available sequences. We studied how genetic variation has changed through time in this subspecies and how it is phylogenetically related to other ringed seal populations from the Baltic Sea, Lake Ladoga, North America, Svalbard, and the White Sea. We observed temporal fluctuations in haplotype frequencies and loss of haplotypes accompanied by a recent reduction in female effective population size. In apparent contrast with the traditionally held view of the Baltic origin of the population, the Saimaa ringed seal mtDNA variation also shows affinities to North American ringed seals. Our results suggest that the Saimaa ringed seal has experienced recent genetic drift associated with small population size. The results further suggest that extant Baltic ringed seal is not representative of the ancestral population of the Saimaa ringed seal, which calls for re-evaluation of the deep history of this subspecies.
Keywords: Saimaa ringed seal; freshwater pinniped; genetic diversity; genetic drift; mitochondrial DNA; museum specimens.
© 2023 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Asplund, T. , Veselov, A. , Primmer, C. R. , Bakhmet, I. , Potutkin, A. , Titov, S. , & Lumme, J. (2004). Geographical structure and postglacial history of mtDNA haplotype variation in Atlantic salmon (Salmo salar L.) among rivers of the white and Barents Sea basins. Annales Zoologici Fennici, 41(3), 465–475.
-
- Baele, G. , Lemey, P. , Bedford, T. , Rambaut, A. , Suchard, M. A. , & Alekseyenko, A. V. (2012). Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Molecular Biology and Evolution, 29(9), 2157–2167. 10.1093/molbev/mss084 - DOI - PMC - PubMed
-
- Berta, A. , & Churchill, M. (2012). Pinniped taxonomy: Review of currently recognized species and subspecies, and evidence used for their description. Mammal Review, 42(3), 207–234. 10.1111/j.1365-2907.2011.00193.x - DOI
-
- Bi, K. , Linderoth, T. , Singhal, S. , Vanderpool, D. , Patton, J. L. , Nielsen, R. , Moritz, C. , & Good, J. M. (2019). Temporal genomic contrasts reveal rapid evolutionary responses in an alpine mammal during recent climate change. PLoS Genetics, 15(5), e1008119. 10.1371/journal.pgen.1008119 - DOI - PMC - PubMed
-
- Borchsenius, F. (2009). FastGap 1.2. http://www.aubot.dk/FastGap_home.htm
LinkOut - more resources
Full Text Sources

