Comparison of in situ ruminal straw fiber degradation and bacterial community between buffalo and Holstein fed with high-roughage diet
- PMID: 36699590
- PMCID: PMC9868309
- DOI: 10.3389/fmicb.2022.1079056
Comparison of in situ ruminal straw fiber degradation and bacterial community between buffalo and Holstein fed with high-roughage diet
Abstract
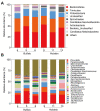
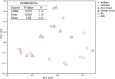
Buffalo exhibits great efficiency in utilizing low-quality roughage, which can be due to the combined effect of host physiological feature and roughage diet fed. The present study was designed to compare the ruminal fiber degradation and the bacterial community attached to straws in buffalo and Holstein when fed with the same high-roughage diet using in situ ruminal incubation technique. Rice and wheat straws were selected as the incubation substrates and sampled at 0, 4, 12, 24, 48, 72, 120, and 216 h of incubation time to measure the kinetics of dry matter (DM) and neutral detergent fiber (NDF) disappearance. Additional two bags were incubated and sampled at 4 and 48 h of incubation time to evaluate the bacterial community attached to straws. The results showed that buffalo exhibited a greater (p ≤ 0.05) fraction of rapidly soluble and washout nutrients and effective ruminal disappearance for both DM and NDF of straw than Holstein, together with a greater (p ≤ 0.05) disappearance rate of potentially degradable nutrient fraction for NDF. Principal coordinate analysis indicated that both host and incubation time altered the bacterial communities attached to straws. Buffalo exhibited greater (p ≤ 0.05) 16S rRNA gene copies of bacteria and greater (p ≤ 0.05) relative abundance of Ruminococcus attached to straw than Holstein. Prolonging incubation time increased (p ≤ 0.05) the 16S rRNA gene copies of bacteria, and the relative abundance of phyla Proteobacteria and Fibrobacters by comparing 4 vs. 48 h of incubation time. In summary, buffalo exhibits greater ruminal fiber degradation than Holstein through increasing bacterial population and enriching Ruminococcus, while prolonging incubation time facilitates fiber degradation through enriching phyla Proteobacteria and Fibrobacteres.
Keywords: bacterial community; buffalo; in situ; rumen ecosystem; straw fiber degradation.
Copyright © 2023 Pu, Zhang, Li, Wang, Zhang, Zhang, Lin, Tan, Tan and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
3-Nitrooxypropanol supplementation had little effect on fiber degradation and microbial colonization of forage particles when evaluated using the in situ ruminal incubation technique.J Dairy Sci. 2020 Oct;103(10):8986-8997. doi: 10.3168/jds.2019-18077. Epub 2020 Aug 26. J Dairy Sci. 2020. PMID: 32861497
-
Brittle Culm 15 mutation alters carbohydrate composition, degradation and methanogenesis of rice straw during in vitro ruminal fermentation.Front Plant Sci. 2022 Aug 5;13:975456. doi: 10.3389/fpls.2022.975456. eCollection 2022. Front Plant Sci. 2022. PMID: 35991441 Free PMC article.
-
Comparison of Ruminal Degradability, Indigestible Neutral Detergent Fiber, and Total-Tract Digestibility of Three Main Crop Straws with Alfalfa Hay and Corn Silage.Animals (Basel). 2021 Nov 11;11(11):3218. doi: 10.3390/ani11113218. Animals (Basel). 2021. PMID: 34827950 Free PMC article.
-
Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants.J Dairy Sci. 2016 Dec;99(12):9668-9681. doi: 10.3168/jds.2016-11398. Epub 2016 Sep 28. J Dairy Sci. 2016. PMID: 27692708
-
Substitution of wheat straw with sugarcane bagasse in low-forage diets fed to mid-lactation dairy cows: Milk production, digestibility, and chewing behavior.J Dairy Sci. 2020 Sep;103(9):8034-8047. doi: 10.3168/jds.2020-18499. Epub 2020 Jul 16. J Dairy Sci. 2020. PMID: 32684450
Cited by
-
Metagenomic sequencing reveals the taxonomic and functional characteristics of rumen microorganisms in Dongliu buffalo.Sci Rep. 2025 May 26;15(1):18398. doi: 10.1038/s41598-025-03059-8. Sci Rep. 2025. PMID: 40419614 Free PMC article.
-
From the perspective of rumen microbiome and host metabolome, revealing the effects of feeding strategies on Jersey Cows on the Tibetan Plateau.PeerJ. 2023 Sep 11;11:e16010. doi: 10.7717/peerj.16010. eCollection 2023. PeerJ. 2023. PMID: 37719116 Free PMC article.
-
Analysis of Rumen Degradation Characteristics, Attached Microbial Community, and Cellulase Activity Changes of Garlic Skin and Artemisia argyi Stalk.Animals (Basel). 2024 Jan 4;14(1):169. doi: 10.3390/ani14010169. Animals (Basel). 2024. PMID: 38200900 Free PMC article.
-
A lysing polysaccharide monooxygenase from Aspergillus niger effectively facilitated rumen microbial fermentation of rice straw.Anim Biosci. 2024 Oct;37(10):1738-1750. doi: 10.5713/ab.24.0026. Epub 2024 May 7. Anim Biosci. 2024. PMID: 38754847 Free PMC article.
-
Mixed Ensiling Increases Degradation Without Altering Attached Microbiota Through In Situ Ruminal Incubation Technique.Animals (Basel). 2025 Jul 18;15(14):2131. doi: 10.3390/ani15142131. Animals (Basel). 2025. PMID: 40723594 Free PMC article.
References
-
- Association of Official Analytical Chemists (AOAC) (2005). Official Methods of Analysis, 18th. AOAC International, Gaithersburg, MD.
-
- Bray J. R., Curtis J. T. (1957). An Ordination of Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268 - DOI
LinkOut - more resources
Full Text Sources

