Microspore embryogenesis induction by mannitol and TSA results in a complex regulation of epigenetic dynamics and gene expression in bread wheat
- PMID: 36699843
- PMCID: PMC9868772
- DOI: 10.3389/fpls.2022.1058421
Microspore embryogenesis induction by mannitol and TSA results in a complex regulation of epigenetic dynamics and gene expression in bread wheat
Abstract
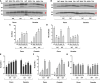
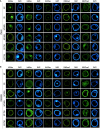
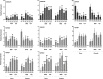
Reprogramming of microspores development towards embryogenesis mediated by stress treatment constitutes the basis of doubled haploid production. Recently, compounds that alter histone post-translational modifications (PTMs) have been reported to enhance microspore embryogenesis (ME), by altering histones acetylation or methylation. However, epigenetic mechanisms underlying ME induction efficiency are poorly understood. In this study, the epigenetic dynamics and the expression of genes associated with histone PTMs and ME induction were studied in two bread wheat cultivars with different ME response. Microspores isolated at 0, 3 and 5 days, treated with 0.7M mannitol (MAN) and 0.7M mannitol plus 0.4µM trichostatin A (TSA), which induced ME more efficiently, were analyzed. An additional control of gametophytic development was included. Microspores epigenetic state at the onset of ME induction was distinctive between cultivars by the ratio of H3 variants and their acetylated forms, the localization and percentage of labeled microspores with H3K9ac, H4K5ac, H4K16ac, H3K9me2 and H3K27me3, and the expression of genes related to pollen development. These results indicated that microspores of the high responding cultivar could be at a less advanced stage in pollen development. MAN and TSA resulted in a hyperacetylation of H3.2, with a greater effect of TSA. Histone PTMs were differentially affected by both treatments, with acetylation being most concerned. The effect of TSA was observed in the H4K5ac localization pattern at 3dT in the mid-low responding cultivar. Three gene networks linked to ME response were identified. TaHDT1, TaHAG2, TaYAO, TaNFD6-A, TabZIPF1 and TaAGO802-B, associated with pollen development, were down-regulated. TaHDA15, TaHAG3, TaHAM, TaYUC11D, Ta-2B-LBD16 TaMS1 and TaDRM3 constituted a network implicated in morphological changes by auxin signaling and cell wall modification up-regulated at 3dT. The last network included TaHDA18, TaHAC1, TaHAC4, TaABI5, TaATG18fD, TaSDG1a-7A and was related to ABA and ethylene hormone signaling pathways, DNA methylation and autophagy processes, reaching the highest expression at 5dT. The results indicated that TSA mainly modified the regulation of genes related to pollen and auxin signaling. This study represents a breakthrough in identifying the epigenetic dynamics and the molecular mechanisms governing ME induction efficiency, with relevance to recalcitrant wheat genotypes and other crops.
Keywords: bread wheat; epigenetics; gene expression; histones; mannitol; microspore embryogenesis; trichostatin A.
Copyright © 2023 Valero-Rubira, Castillo, Burrell and Vallés.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
New Epigenetic Modifier Inhibitors Enhance Microspore Embryogenesis in Bread Wheat.Plants (Basel). 2024 Mar 8;13(6):772. doi: 10.3390/plants13060772. Plants (Basel). 2024. PMID: 38592809 Free PMC article.
-
Trichostatin A Affects Developmental Reprogramming of Bread Wheat Microspores towards an Embryogenic Route.Plants (Basel). 2020 Oct 26;9(11):1442. doi: 10.3390/plants9111442. Plants (Basel). 2020. PMID: 33114625 Free PMC article.
-
Increase of histone acetylation by suberoylanilide hydroxamic acid enhances microspore reprogramming and expression of somatic embryogenesis transcription factors in Brassica napus.Plant Sci. 2025 Feb;351:112318. doi: 10.1016/j.plantsci.2024.112318. Epub 2024 Nov 20. Plant Sci. 2025. PMID: 39577504
-
Microspore embryogenesis: targeting the determinant factors of stress-induced cell reprogramming for crop improvement.J Exp Bot. 2019 Jun 1;70(11):2965-2978. doi: 10.1093/jxb/ery464. J Exp Bot. 2019. PMID: 30753698 Review.
-
Microspore embryogenesis in Brassica: calcium signaling, epigenetic modification, and programmed cell death.Planta. 2018 Dec;248(6):1339-1350. doi: 10.1007/s00425-018-2996-5. Epub 2018 Aug 31. Planta. 2018. PMID: 30171331 Review.
Cited by
-
New Epigenetic Modifier Inhibitors Enhance Microspore Embryogenesis in Bread Wheat.Plants (Basel). 2024 Mar 8;13(6):772. doi: 10.3390/plants13060772. Plants (Basel). 2024. PMID: 38592809 Free PMC article.
-
Cu2+ and Zn2+ Ions Affecting Biochemical Paths and DNA Methylation of Rye (Secale cereale L.) Anther Culture Influencing Plant Regeneration Efficiency.Cells. 2025 Jul 29;14(15):1167. doi: 10.3390/cells14151167. Cells. 2025. PMID: 40801598 Free PMC article.
-
Plant Growth Regulation in Cell and Tissue Culture In Vitro.Plants (Basel). 2024 Jan 22;13(2):327. doi: 10.3390/plants13020327. Plants (Basel). 2024. PMID: 38276784 Free PMC article. Review.
-
The improvement of the in vitro plant regeneration in barley with the epigenetic modifier of histone acetylation, trichostatin A.J Appl Genet. 2024 Feb;65(1):13-30. doi: 10.1007/s13353-023-00800-9. Epub 2023 Nov 14. J Appl Genet. 2024. PMID: 37962803 Free PMC article.
-
Microinjection of the CRISPR/Cas9 editing system through the germ pore of a wheat microspore induces mutations in the target Ms2 gene.Mol Biol Rep. 2024 Jun 1;51(1):706. doi: 10.1007/s11033-024-09644-w. Mol Biol Rep. 2024. PMID: 38824203
References
-
- Akond Z., Rahman H., Ahsan M. A., Mosharaf M. P., Alam M., Mollah M. N. H. (2020). Bioinformatic analysis based genome-wide identification, characterization, diversification and regulatory transcription components of RNA silencing machinery genes in wheat (Triticum aestivum l.). bioRxiv. doi: 10.1101/2020.05.21.108100 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

