Starting from scratch: Step-by-step development of diagnostic tests for SARS-CoV-2 detection by RT-LAMP
- PMID: 36701313
- PMCID: PMC9879405
- DOI: 10.1371/journal.pone.0279681
Starting from scratch: Step-by-step development of diagnostic tests for SARS-CoV-2 detection by RT-LAMP
Abstract
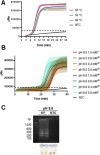
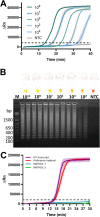
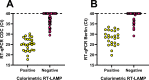
The pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected millions of people worldwide. Public health strategies to reduce viral transmission are based on widespread diagnostic testing to detect and isolate contagious patients. Several reverse transcription (RT)-PCR tests, along with other SARS-CoV-2 diagnostic assays, are available to attempt to cover the global demand. Loop-mediated isothermal amplification (LAMP) based methods have been established as rapid, accurate, point of care diagnostic tests for viral infections; hence, they represent an excellent alternative for SARS-CoV-2 detection. The aim of this study was to develop and describe molecular detection systems for SARS-CoV-2 based on RT-LAMP. Recombinant DNA polymerase from Bacillus stearothermophilus and thermostable engineered reverse transcriptase from Moloney Murine Leukemia Virus were expressed using a prokaryotic system and purified by fast protein liquid chromatography. These enzymes were used to set up fluorometric real time and colorimetric end-point RT-LAMP assays. Several reaction conditions were optimized such as reaction temperature, Tris-HCl concentration, and pH of the diagnostic tests. The key enzymes for RT-LAMP were purified and their enzymatic activity was determined. Standardized reaction conditions for both RT-LAMP assays were 65°C and a Tris-HCl-free buffer at pH 8.8. Colorimetric end-point RT-LAMP assay was successfully used for viral detection from clinical saliva samples with 100% sensitivity and 100% specificity compared to the results obtained by RT-qPCR based diagnostic protocols with Ct values until 30. The developed RT-LAMP diagnostic tests based on purified recombinant enzymes allowed a sensitive and specific detection of the nucleocapsid gene of SARS-CoV-2.
Copyright: © 2023 Tapia-Sidas et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: The detection using RT-LAMP was registered by Centro de Investigación y de Estudios Avanzados (CINVESTAV) to be protected as a patent. Biopure® is a registered trademark of CINVESTAV. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures

Similar articles
-
A Multiplex and Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Sensitive and Rapid Detection of Novel SARS-CoV-2.Front Cell Infect Microbiol. 2021 Jun 29;11:653616. doi: 10.3389/fcimb.2021.653616. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34268131 Free PMC article.
-
Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification.Clin Chem. 2021 Jan 30;67(2):415-424. doi: 10.1093/clinchem/hvaa267. Clin Chem. 2021. PMID: 33098427 Free PMC article.
-
A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples.Sci Transl Med. 2020 Aug 12;12(556):eabc7075. doi: 10.1126/scitranslmed.abc7075. Epub 2020 Jul 27. Sci Transl Med. 2020. PMID: 32719001 Free PMC article.
-
A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview.Front Immunol. 2021 Dec 14;12:732756. doi: 10.3389/fimmu.2021.732756. eCollection 2021. Front Immunol. 2021. PMID: 34970254 Free PMC article. Review.
-
Advances in RT-LAMP for COVID-19 testing and diagnosis.Expert Rev Mol Diagn. 2023 Jan;23(1):9-28. doi: 10.1080/14737159.2023.2169071. Epub 2023 Mar 1. Expert Rev Mol Diagn. 2023. PMID: 36695788 Review.
Cited by
-
Quantitative mRNA expression measurement at home.Sci Rep. 2024 Jan 10;14(1):1013. doi: 10.1038/s41598-023-49651-8. Sci Rep. 2024. PMID: 38200031 Free PMC article.
-
A Comparative Analysis of Molecular Biological Methods for the Detection of SARS-CoV-2 and Testing the In Vitro Infectivity of the Virus.Microorganisms. 2024 Jan 17;12(1):180. doi: 10.3390/microorganisms12010180. Microorganisms. 2024. PMID: 38258006 Free PMC article.
-
Development of a rapid LFA test based on direct RT-LAMP for diagnosis of SARS-CoV-2.Pract Lab Med. 2024 Oct 18;42:e00437. doi: 10.1016/j.plabm.2024.e00437. eCollection 2024 Nov. Pract Lab Med. 2024. PMID: 39553462 Free PMC article.
-
Laboratory Evaluation of a SARS-CoV-2 RT-LAMP Test.Trop Med Infect Dis. 2023 Jun 13;8(6):320. doi: 10.3390/tropicalmed8060320. Trop Med Infect Dis. 2023. PMID: 37368738 Free PMC article.
References
-
- WHO. Rolling updates on coronavirus disease (COVID-19). World Health Organ 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-a....
-
- CDC. Coronavirus Disease 2019 (COVID-19): Interim Clinical Guidance for Management of Patients with Confirmed Coronavirus Disease (COVID-19). Cent Dis Control Prev 2020. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-manageme....
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

