A Distributional Model of Bound Ligand Conformational Strain: From Small Molecules up to Large Peptidic Macrocycles
- PMID: 36701387
- PMCID: PMC9923749
- DOI: 10.1021/acs.jmedchem.2c01744
A Distributional Model of Bound Ligand Conformational Strain: From Small Molecules up to Large Peptidic Macrocycles
Abstract
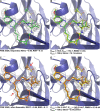
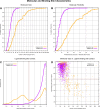
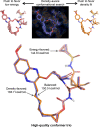
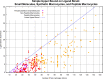
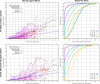
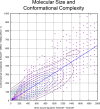
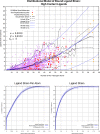
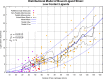
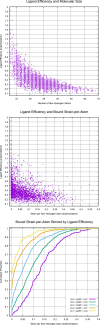
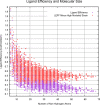
The internal conformational strain incurred by ligands upon binding a target site has a critical impact on binding affinity, and expectations about the magnitude of ligand strain guide conformational search protocols. Estimates for bound ligand strain begin with modeled ligand atomic coordinates from X-ray co-crystal structures. By deriving low-energy conformational ensembles to fit X-ray diffraction data, calculated strain energies are substantially reduced compared with prior approaches. We show that the distribution of expected global strain energy values is dependent on molecular size in a superlinear manner. The distribution of strain energy follows a rectified normal distribution whose mean and variance are related to conformational complexity. The modeled strain distribution closely matches calculated strain values from experimental data comprising over 3000 protein-ligand complexes. The distributional model has direct implications for conformational search protocols as well as for directions in molecular design.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

