Characterization of SpsQ from Staphylococcus pseudintermedius as an affinity chromatography ligand for canine therapeutic antibodies
- PMID: 36701408
- PMCID: PMC9879442
- DOI: 10.1371/journal.pone.0281171
Characterization of SpsQ from Staphylococcus pseudintermedius as an affinity chromatography ligand for canine therapeutic antibodies
Abstract
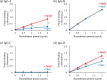
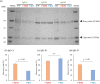
Coagulase-positive Staphylococci express protein A, which binds to host antibodies, to evade the immune system. Taking advantage of its specific binding to antibodies, protein A from Staphylococcus aureus, which is called SpA, is commonly used as an affinity chromatography ligand for human therapeutic antibodies. However, among four canine IgG subclasses (A, B, C, and D), only IgG-B binds to SpA strongly and establishing an efficient and robust purification scheme for canine therapeutic antibodies whose IgG subclass is A, C, or D remains difficult and depends on finding a suitable substitute to SpA. S. pseudintermedius, a major coagulase-positive Staphylococci found in dogs, expresses spsQ gene which is orthologous to S. aureus spa. We hypothesized that to serve S. pseudintermedius to better adapt to the dog immune system, SpsQ would bind to canine IgGs stronger than SpA, making it a better affinity chromatography ligand for canine therapeutic antibodies. To characterize SpsQ, we first determined the spsQ nucleotide sequence from S. pseudintermedius isolates. Based on the identified sequence, we prepared recombinant proteins containing the immunoglobulin-binding domains of SpA (r-SpA) and SpsQ (r-SpsQ) and determined their binding capacity for each canine IgG subclass. The binding capacity of r-SpsQ for IgG-B was almost as high as that of r-SpA. Interestingly, while both r-SpsQ and r-SpA showed no binding to IgG-C, the binding capacity of r-SpsQ for IgG-A and IgG-D was significantly higher than that of r-SpA. Finally, we performed affinity chromatography using r-SpsQ- or r-SpA-immobilized resin and revealed that the recovery rates of IgG-A and IgG-D using r-SpsQ were significantly higher than those using r-SpA. Our findings indicate that SpsQ has a strong potential to be used as an affinity chromatography ligand for canine therapeutic antibodies of subclass A, B, and D.
Copyright: © 2023 Takeuchi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Characterization of recombinant wild-type and nontoxigenic protein A from Staphylococcus pseudintermedius.Virulence. 2018;9(1):1050-1061. doi: 10.1080/21505594.2018.1489199. Virulence. 2018. PMID: 30052123 Free PMC article.
-
Staphylococcal protein A binding to canine IgG and IgM.Vet Immunol Immunopathol. 1997 Nov;59(3-4):205-12. doi: 10.1016/s0165-2427(97)00073-1. Vet Immunol Immunopathol. 1997. PMID: 9477472
-
Expression and function of protein A in Staphylococcus pseudintermedius.Virulence. 2018 Jan 1;9(1):390-401. doi: 10.1080/21505594.2017.1403710. Virulence. 2018. PMID: 29157101 Free PMC article.
-
Immunoglobulin purification by affinity chromatography using protein A mimetic ligands prepared by combinatorial chemical synthesis.Immunol Invest. 2002 Aug-Nov;31(3-4):263-78. doi: 10.1081/imm-120016245. Immunol Invest. 2002. PMID: 12472184 Review.
-
A comprehensive review on staphylococcal protein A (SpA): Its production and applications.Biotechnol Appl Biochem. 2019 May;66(3):454-464. doi: 10.1002/bab.1742. Epub 2019 Mar 21. Biotechnol Appl Biochem. 2019. PMID: 30869160 Review.
Cited by
-
[Preparation technology comparison and performance evaluation of different protein A affinity chromatographic materials].Se Pu. 2024 Apr 8;42(5):410-419. doi: 10.3724/SP.J.1123.2024.01018. Se Pu. 2024. PMID: 38736384 Free PMC article. Chinese.
-
Virulence Mechanisms of Staphylococcal Animal Pathogens.Int J Mol Sci. 2023 Sep 26;24(19):14587. doi: 10.3390/ijms241914587. Int J Mol Sci. 2023. PMID: 37834035 Free PMC article. Review.
References
-
- Forsgren A, Sjöquist J. “Protein A” from S. Aureus: I. Pseudo-Immune Reaction with Human γ -Globulin. J Immunol 1966; 97:822–827. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

