Evolution of giant pandoravirus revealed by CRISPR/Cas9
- PMID: 36702819
- PMCID: PMC9879987
- DOI: 10.1038/s41467-023-36145-4
Evolution of giant pandoravirus revealed by CRISPR/Cas9
Abstract
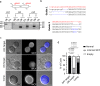
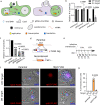
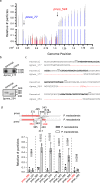
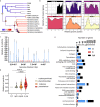
Giant viruses (GVs) are a hotspot of unresolved controversies since their discovery, including the definition of "Virus" and their origin. While increasing knowledge of genome diversity has accumulated, GV functional genomics was largely neglected. Here, we describe an experimental framework to genetically modify nuclear GVs and their host Acanthamoeba castellanii using CRISPR/Cas9, shedding light on the evolution from small icosahedral viruses to amphora-shaped GVs. Ablation of the icosahedral major capsid protein in the phylogenetically-related mollivirus highlights a transition in virion shape and size. We additionally demonstrate the existence of a reduced core essential genome in pandoravirus, reminiscent of their proposed smaller ancestors. This proposed genetic expansion led to increased genome robustness, indicating selective pressures for adaptation to uncertain environments. Overall, we introduce new tools for manipulation of the unexplored genome of nuclear GVs and provide experimental evidence suggesting that viral gigantism has aroused as an emerging trait.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

