Dermal macrophages set pain sensitivity by modulating the amount of tissue NGF through an SNX25-Nrf2 pathway
- PMID: 36703006
- PMCID: PMC9977679
- DOI: 10.1038/s41590-022-01418-5
Dermal macrophages set pain sensitivity by modulating the amount of tissue NGF through an SNX25-Nrf2 pathway
Abstract
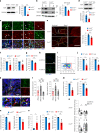
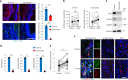
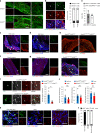
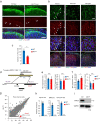
Cross-talk between peripheral neurons and immune cells is important in pain sensation. We identified Snx25 as a pain-modulating gene in a transgenic mouse line with reduced pain sensitivity. Conditional deletion of Snx25 in monocytes and macrophages, but not in peripheral sensory neurons, in mice (Snx25cKO mice) reduced pain responses in both normal and neuropathic conditions. Bone marrow transplantation using Snx25cKO and wild-type mice indicated that macrophages modulated pain sensitivity. Expression of sorting nexin (SNX)25 in dermal macrophages enhanced expression of the neurotrophic factor NGF through the inhibition of ubiquitin-mediated degradation of Nrf2, a transcription factor that activates transcription of Ngf. As such, dermal macrophages set the threshold for pain sensitivity through the production and secretion of NGF into the dermis, and they may cooperate with dorsal root ganglion macrophages in pain perception.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Macrophages set the bar for acute pain sensitivity.Nat Immunol. 2023 Mar;24(3):382-384. doi: 10.1038/s41590-023-01438-9. Nat Immunol. 2023. PMID: 36823407 No abstract available.
Similar articles
-
Dermal macrophages control tactile perception under physiological conditions via NGF signaling.Sci Rep. 2024 Nov 8;14(1):27192. doi: 10.1038/s41598-024-78683-x. Sci Rep. 2024. PMID: 39516548 Free PMC article.
-
SNX25 regulates proinflammatory cytokine expression via the NF-κB signal in macrophages.PLoS One. 2021 Mar 1;16(3):e0247840. doi: 10.1371/journal.pone.0247840. eCollection 2021. PLoS One. 2021. PMID: 33647065 Free PMC article.
-
Nerve growth factor acts through the TrkA receptor to protect sensory neurons from the damaging effects of the HIV-1 viral protein, Vpr.Neuroscience. 2013 Nov 12;252:512-25. doi: 10.1016/j.neuroscience.2013.07.046. Epub 2013 Jul 30. Neuroscience. 2013. PMID: 23912036 Free PMC article.
-
Role of the immune system in neuropathic pain.Scand J Pain. 2019 Dec 18;20(1):33-37. doi: 10.1515/sjpain-2019-0138. Scand J Pain. 2019. PMID: 31730538 Review.
-
Phenotypic modification of primary sensory neurons: the role of nerve growth factor in the production of persistent pain.Philos Trans R Soc Lond B Biol Sci. 1996 Mar 29;351(1338):441-8. doi: 10.1098/rstb.1996.0040. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8730783 Review.
Cited by
-
Cancer Therapy-induced Dermatotoxicity as a Window to Understanding Skin Immunity.Hematol Oncol Clin North Am. 2024 Oct;38(5):1011-1025. doi: 10.1016/j.hoc.2024.05.002. Epub 2024 Jun 12. Hematol Oncol Clin North Am. 2024. PMID: 38866636 Review.
-
Catalytic neural stem cell exosomes for multi-stage targeting and synergistical therapy of retinal ischemia-reperfusion injury.Cell Rep Med. 2025 Apr 15;6(4):102052. doi: 10.1016/j.xcrm.2025.102052. Cell Rep Med. 2025. PMID: 40239632 Free PMC article.
-
Pathogenic Crosstalk Between the Peripheral and Central Nervous System in Rheumatic Diseases: Emerging Evidence and Clinical Implications.Int J Mol Sci. 2025 Jun 24;26(13):6036. doi: 10.3390/ijms26136036. Int J Mol Sci. 2025. PMID: 40649815 Free PMC article. Review.
-
Assessing the Causal Relationship Between Immune Cells and Temporomandibular Related Pain by Bi‑Directional Mendelian Randomization Analysis.J Pain Res. 2024 Nov 16;17:3791-3800. doi: 10.2147/JPR.S486817. eCollection 2024. J Pain Res. 2024. PMID: 39574830 Free PMC article.
-
Immunotherapies in chronic pain through modulation of neuroimmune interactions.Pharmacol Ther. 2023 Aug;248:108476. doi: 10.1016/j.pharmthera.2023.108476. Epub 2023 Jun 10. Pharmacol Ther. 2023. PMID: 37307899 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

