Reading mixtures of uniform sequence-defined macromolecules to increase data storage capacity
- PMID: 36703345
- PMCID: PMC9814948
- DOI: 10.1038/s42004-020-00431-9
Reading mixtures of uniform sequence-defined macromolecules to increase data storage capacity
Abstract
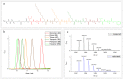
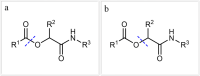
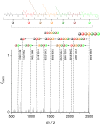
In recent years, the field of molecular data storage has emerged from a niche to a vibrant research topic. Herein, we describe a simultaneous and automated read-out of data stored in mixtures of sequence-defined oligomers. Therefore, twelve different sequence-defined tetramers and three hexamers with different mass markers and side chains are successfully synthesised via iterative Passerini three-component reactions and subsequent deprotection steps. By programming a straightforward python script for ESI-MS/MS analysis, it is possible to automatically sequence and thus read-out the information stored in these oligomers within one second. Most importantly, we demonstrate that the use of mass-markers as starting compounds eases MS/MS data interpretation and furthermore allows the unambiguous reading of sequences of mixtures of sequence-defined oligomers. Thus, high data storage capacity considering the field of synthetic macromolecules (up to 64.5 bit in our examples) can be obtained without the need of synthesizing long sequences, but by mixing and simultaneously analysing shorter sequence-defined oligomers.
© 2020. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Dual sequence definition increases the data storage capacity of sequence-defined macromolecules.Commun Chem. 2020 May 20;3(1):63. doi: 10.1038/s42004-020-0308-z. Commun Chem. 2020. PMID: 36703457 Free PMC article.
-
A Scalable and High-Yield Strategy for the Synthesis of Sequence-Defined Macromolecules.Angew Chem Int Ed Engl. 2016 Jan 18;55(3):1204-7. doi: 10.1002/anie.201509398. Epub 2015 Dec 9. Angew Chem Int Ed Engl. 2016. PMID: 26663541
-
Rewritable Macromolecular Data Storage with Automated Read-out.Angew Chem Int Ed Engl. 2022 Mar 21;61(13):e202116718. doi: 10.1002/anie.202116718. Epub 2022 Feb 11. Angew Chem Int Ed Engl. 2022. PMID: 35104375
-
Reading Information Stored in Synthetic Macromolecules.J Am Chem Soc. 2022 Dec 14;144(49):22378-22390. doi: 10.1021/jacs.2c10316. Epub 2022 Dec 1. J Am Chem Soc. 2022. PMID: 36454647 Review.
-
Recent Progress in the Design of Monodisperse, Sequence-Defined Macromolecules.Macromol Rapid Commun. 2017 May;38(9). doi: 10.1002/marc.201600711. Epub 2017 Mar 15. Macromol Rapid Commun. 2017. PMID: 28297122 Review.
Cited by
-
Molecular Encryption and Steganography Using Mixtures of Simultaneously Sequenced, Sequence-Defined Oligourethanes.ACS Cent Sci. 2022 Aug 24;8(8):1125-1133. doi: 10.1021/acscentsci.2c00460. Epub 2022 Jul 20. ACS Cent Sci. 2022. PMID: 36032764 Free PMC article.
-
Molecular data storage using direct analysis in real time (DART) ionization mass spectrometry for decoding.Sci Rep. 2023 Oct 3;13(1):16576. doi: 10.1038/s41598-023-43658-x. Sci Rep. 2023. PMID: 37789061 Free PMC article.
-
Molecular data storage with zero synthetic effort and simple read-out.Sci Rep. 2022 Aug 16;12(1):13878. doi: 10.1038/s41598-022-18108-9. Sci Rep. 2022. PMID: 35974033 Free PMC article.
-
Complex Sequence-Defined Heteropolymers Enable Controlled Film Growth in Layer-By-Layer Assembly.Macromol Rapid Commun. 2024 Nov;45(22):e2400482. doi: 10.1002/marc.202400482. Epub 2024 Aug 6. Macromol Rapid Commun. 2024. PMID: 39108056 Free PMC article.
References
-
- Jenkins AD, Stepto RFT, Kratochvíl P, Suter UW. Glossary of basic terms in polymer science. Pure Appl. Chem. 1996;68:2287–2311. doi: 10.1351/pac199668122287. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

