Navigating the DNA encoded libraries chemical space
- PMID: 36703354
- PMCID: PMC9814293
- DOI: 10.1038/s42004-020-00374-1
Navigating the DNA encoded libraries chemical space
Abstract
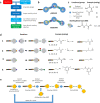
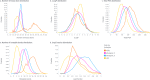
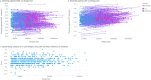
DNA-encoded library (DEL) technology is a novel ligand identification strategy that allows the synthesis and screening of unprecedented chemical diversity more efficiently than conventional methods. However, no reports have been published to systematically study how to increase the diversity and improve the molecular property space that can be covered with DEL. This report describes the development and application of eDESIGNER, an algorithm that comprehensively generates all possible library designs, enumerates and profiles samples from each library and evaluates them to select the libraries to be synthesized. This tool utilizes suitable on-DNA chemistries and available building blocks to design and identify libraries with a pre-defined molecular weight distribution and maximal diversity compared with compound collections from other sources.
© 2020. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
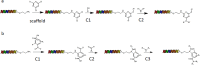

Similar articles
-
Exploration of the Chemical Space of DNA-encoded Libraries.Mol Inform. 2022 Jun;41(6):e2100289. doi: 10.1002/minf.202100289. Epub 2022 Jan 28. Mol Inform. 2022. PMID: 34981643
-
DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries.Acc Chem Res. 2014 Apr 15;47(4):1247-55. doi: 10.1021/ar400284t. Epub 2014 Mar 28. Acc Chem Res. 2014. PMID: 24673190
-
Diversity-oriented synthesis as a tool to expand the chemical space of DNA-encoded libraries.Bioorg Med Chem. 2021 Jul 1;41:116218. doi: 10.1016/j.bmc.2021.116218. Epub 2021 May 15. Bioorg Med Chem. 2021. PMID: 34030087 Review.
-
DNA-encoded libraries (DELs): a review of on-DNA chemistries and their output.RSC Adv. 2021 Jan 19;11(4):2359-2376. doi: 10.1039/d0ra09889b. eCollection 2021 Jan 6. RSC Adv. 2021. PMID: 35424149 Free PMC article. Review.
-
Evolution of the Selection Methods of DNA-Encoded Chemical Libraries.Acc Chem Res. 2021 Sep 7;54(17):3491-3503. doi: 10.1021/acs.accounts.1c00375. Epub 2021 Aug 24. Acc Chem Res. 2021. PMID: 34427078
Cited by
-
The Growing Importance of Chirality in 3D Chemical Space Exploration and Modern Drug Discovery Approaches for Hit-ID: Topical Innovations.ACS Med Chem Lett. 2021 Jul 16;12(8):1220-1229. doi: 10.1021/acsmedchemlett.1c00251. eCollection 2021 Aug 12. ACS Med Chem Lett. 2021. PMID: 34413951 Free PMC article.
-
Small Molecule Modulator of the mTORC2 Pathway Discovered from a DEL Library Designed to Bind to Pleckstrin Homology Domains.ACS Chem Biol. 2024 Dec 20;19(12):2502-2514. doi: 10.1021/acschembio.4c00597. Epub 2024 Nov 12. ACS Chem Biol. 2024. PMID: 39530383 Free PMC article.
-
Tunable Aziridinium Ylide Reactivity: Non-covalent Interactions Enable Divergent Product Outcomes.ACS Catal. 2022 Jan 21;12(2):1572-1580. doi: 10.1021/acscatal.1c05413. Epub 2022 Jan 11. ACS Catal. 2022. PMID: 35291380 Free PMC article.
-
Navigating chemical reaction space - application to DNA-encoded chemistry.Chem Sci. 2022 Sep 1;13(37):11221-11231. doi: 10.1039/d2sc02474h. eCollection 2022 Sep 28. Chem Sci. 2022. PMID: 36320474 Free PMC article.
-
Development of the Inverse Sonogashira Reaction for DEL Synthesis.ACS Med Chem Lett. 2023 Feb 23;14(3):270-277. doi: 10.1021/acsmedchemlett.2c00477. eCollection 2023 Mar 9. ACS Med Chem Lett. 2023. PMID: 36923912 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

