Quantification of amyloid fibril polymorphism by nano-morphometry reveals the individuality of filament assembly
- PMID: 36703355
- PMCID: PMC9814634
- DOI: 10.1038/s42004-020-00372-3
Quantification of amyloid fibril polymorphism by nano-morphometry reveals the individuality of filament assembly
Abstract
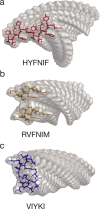
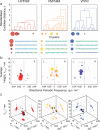
Amyloid fibrils are highly polymorphic structures formed by many different proteins. They provide biological function but also abnormally accumulate in numerous human diseases. The physicochemical principles of amyloid polymorphism are not understood due to lack of structural insights at the single-fibril level. To identify and classify different fibril polymorphs and to quantify the level of heterogeneity is essential to decipher the precise links between amyloid structures and their functional and disease associated properties such as toxicity, strains, propagation and spreading. Employing gentle, force-distance curve-based AFM, we produce detailed images, from which the 3D reconstruction of individual filaments in heterogeneous amyloid samples is achieved. Distinctive fibril polymorphs are then classified by hierarchical clustering, and sample heterogeneity is objectively quantified. These data demonstrate the polymorphic nature of fibril populations, provide important information regarding the energy landscape of amyloid self-assembly, and offer quantitative insights into the structural basis of polymorphism in amyloid populations.
© 2020. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
On the Structural Diversity and Individuality of Polymorphic Amyloid Protein Assemblies.J Mol Biol. 2021 Oct 1;433(20):167124. doi: 10.1016/j.jmb.2021.167124. Epub 2021 Jul 2. J Mol Biol. 2021. PMID: 34224749 Review.
-
Three-dimensional reconstruction of individual helical nano-filament structures from atomic force microscopy topographs.Biomol Concepts. 2020 May 6;11(1):102-115. doi: 10.1515/bmc-2020-0009. Biomol Concepts. 2020. PMID: 32374275
-
Trace_y: Software algorithms for structural analysis of individual helical filaments by three-dimensional contact point reconstruction atomic force microscopy.Structure. 2025 Feb 6;33(2):363-371.e2. doi: 10.1016/j.str.2024.11.007. Epub 2024 Dec 5. Structure. 2025. PMID: 39642871
-
Identifying Polymorphs of Amyloid-β (1-40) Fibrils Using High-Resolution Atomic Force Microscopy.J Phys Chem B. 2019 Dec 12;123(49):10376-10383. doi: 10.1021/acs.jpcb.9b07854. Epub 2019 Dec 4. J Phys Chem B. 2019. PMID: 31714085
-
Molecular rules governing the structural polymorphism of amyloid fibrils in neurodegenerative diseases.Structure. 2023 Nov 2;31(11):1335-1347. doi: 10.1016/j.str.2023.08.006. Epub 2023 Aug 31. Structure. 2023. PMID: 37657437 Review.
Cited by
-
Data-mining unveils structure-property-activity correlation of viral infectivity enhancing self-assembling peptides.Nat Commun. 2023 Aug 23;14(1):5121. doi: 10.1038/s41467-023-40663-6. Nat Commun. 2023. PMID: 37612273 Free PMC article.
-
Direct Observation of Competing Prion Protein Fibril Populations with Distinct Structures and Kinetics.ACS Nano. 2023 Apr 11;17(7):6575-6588. doi: 10.1021/acsnano.2c12009. Epub 2023 Feb 20. ACS Nano. 2023. PMID: 36802500 Free PMC article.
-
A Direct Relationship Between 'Blood Stasis' and Fibrinaloid Microclots in Chronic, Inflammatory, and Vascular Diseases, and Some Traditional Natural Products Approaches to Treatment.Pharmaceuticals (Basel). 2025 May 12;18(5):712. doi: 10.3390/ph18050712. Pharmaceuticals (Basel). 2025. PMID: 40430532 Free PMC article. Review.
-
Generating Ensembles of Dynamic Misfolding Proteins.Front Neurosci. 2022 Mar 31;16:881534. doi: 10.3389/fnins.2022.881534. eCollection 2022. Front Neurosci. 2022. PMID: 35431773 Free PMC article. Review.
-
Comparative Analysis of the Relative Fragmentation Stabilities of Polymorphic Alpha-Synuclein Amyloid Fibrils.Biomolecules. 2022 Apr 25;12(5):630. doi: 10.3390/biom12050630. Biomolecules. 2022. PMID: 35625557 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

