Dual sequence definition increases the data storage capacity of sequence-defined macromolecules
- PMID: 36703457
- PMCID: PMC9814518
- DOI: 10.1038/s42004-020-0308-z
Dual sequence definition increases the data storage capacity of sequence-defined macromolecules
Abstract
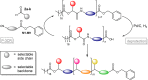
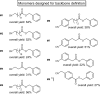
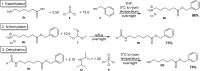
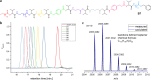
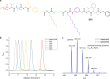
Sequence-defined macromolecules offer applications in the field of data storage. Challenges include synthesising precise and pure sequences, reading stored information and increasing data storage capacity. Herein, the synthesis of dual sequence-defined oligomers and their application for data storage is demonstrated. While applying the well-established Passerini three-component reaction, the degree of definition of the prepared monodisperse macromolecules is improved compared to previous reports by utilising nine specifically designed isocyanide monomers to introduce backbone definition. The monomers are combined with various aldehyde components to synthesise dual-sequence defined oligomers. Thus, the side chains and the backbones of these macromolecules can be varied independently, exhibiting increased molecular diversity and hence data storage capacity per repeat unit. In case of a dual sequence-defined pentamer, 33 bits are achieved in a single molecule. The oligomers are obtained in multigram scale and excellent purity. Sequential read-out by tandem ESI-MS/MS verifies the high data storage capacity of the prepared oligomers per repeat unit in comparison to other sequence defined macromolecules.
© 2020. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Reading mixtures of uniform sequence-defined macromolecules to increase data storage capacity.Commun Chem. 2020 Dec 9;3(1):184. doi: 10.1038/s42004-020-00431-9. Commun Chem. 2020. PMID: 36703345 Free PMC article.
-
A Scalable and High-Yield Strategy for the Synthesis of Sequence-Defined Macromolecules.Angew Chem Int Ed Engl. 2016 Jan 18;55(3):1204-7. doi: 10.1002/anie.201509398. Epub 2015 Dec 9. Angew Chem Int Ed Engl. 2016. PMID: 26663541
-
A Combined Photochemical and Multicomponent Reaction Approach to Precision Oligomers.Chemistry. 2018 Mar 7;24(14):3413-3419. doi: 10.1002/chem.201705939. Epub 2018 Feb 5. Chemistry. 2018. PMID: 29337381
-
Recent Progress in the Design of Monodisperse, Sequence-Defined Macromolecules.Macromol Rapid Commun. 2017 May;38(9). doi: 10.1002/marc.201600711. Epub 2017 Mar 15. Macromol Rapid Commun. 2017. PMID: 28297122 Review.
-
A New Class of Materials: Sequence-Defined Macromolecules and Their Emerging Applications.Adv Mater. 2019 Jun;31(26):e1806027. doi: 10.1002/adma.201806027. Epub 2019 Jan 2. Adv Mater. 2019. PMID: 30600565 Review.
Cited by
-
Molecular data storage with zero synthetic effort and simple read-out.Sci Rep. 2022 Aug 16;12(1):13878. doi: 10.1038/s41598-022-18108-9. Sci Rep. 2022. PMID: 35974033 Free PMC article.
-
Multistate Dihydroazulene-Spiropyran Dyads: Path-Dependent Switchings and Refinement of the "Meta-rule" of Photoactivity.Chemistry. 2025 May 22;31(29):e202501061. doi: 10.1002/chem.202501061. Epub 2025 Apr 27. Chemistry. 2025. PMID: 40197595 Free PMC article.
-
Applications of Discrete Synthetic Macromolecules in Life and Materials Science: Recent and Future Trends.Adv Sci (Weinh). 2021 Jan 25;8(6):2004038. doi: 10.1002/advs.202004038. eCollection 2021 Mar. Adv Sci (Weinh). 2021. PMID: 33747749 Free PMC article. Review.
-
Reading mixtures of uniform sequence-defined macromolecules to increase data storage capacity.Commun Chem. 2020 Dec 9;3(1):184. doi: 10.1038/s42004-020-00431-9. Commun Chem. 2020. PMID: 36703345 Free PMC article.
-
Nondestructive Sequencing of Enantiopure Oligoesters by Nuclear Magnetic Resonance Spectroscopy.JACS Au. 2022 Aug 15;2(9):2108-2118. doi: 10.1021/jacsau.2c00388. eCollection 2022 Sep 26. JACS Au. 2022. PMID: 36186555 Free PMC article.
References
-
- Jenkins, A. D., Kratochvíl, P., Stepto, R. F. T. & Suter, U. W. Polymer divisioncommission on macromolecular nomenclature. Pure Appl. Chem. 68 2287–2311 (1996).
-
- Holloway JO, Wetzel KS, Martens S, Du Prez FE, Meier MAR. Direct comparison of solution and solid phase synthesis of sequence-defined macromolecules. Polym. Chem. 2019;10:3859–3867. doi: 10.1039/C9PY00558G. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

