Identification and validation of a prognostic signature of autophagy, apoptosis and pyroptosis-related genes for head and neck squamous cell carcinoma: to imply therapeutic choices of HPV negative patients
- PMID: 36703967
- PMCID: PMC9872116
- DOI: 10.3389/fimmu.2022.1100417
Identification and validation of a prognostic signature of autophagy, apoptosis and pyroptosis-related genes for head and neck squamous cell carcinoma: to imply therapeutic choices of HPV negative patients
Abstract
Introduction: An effective tool is needed to predict the prognosis of head and neck squamous cell carcinoma (HNSCC). Human papillomavirus (HPV) positive HNSCC patients generally have a favorable survival and a promising responsiveness to radiotherapy, chemoradiotherapy and checkpoint blockades. However, HPV negative patients, the majority of HNSCC patients, have been largely overlooked. Cell death has been involved in the therapeutic resistance of cancers. To this end, we aimed to identify the association of autophagy, apoptosis and pyroptosis-related genes with the prognosis of HNSCC, and construct a prognostic signature to predict the prognosis for HNSCC, especially for HPV negative HNSCC.
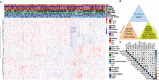
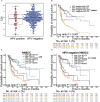
Methods: Autophagy and apoptosis-related genes were obtained from Gene Set Enrichment Analysis (GSEA) website, and pyroptosis-related genes were obtained from GSEA and Gene Ontology (GO) database. We established the cell death index (CDI) based on RNA sequencing (RNA-seq) data and clinicopathological information from The Cancer Genome Atlas (TCGA) dataset. The prognostic value of CDI was verified by Kaplan-Meier, receiver operating characteristic (ROC) and univariate and multivariate Cox regression analyses in TCGA dataset, and validated with the datasets from Gene Expression Omnibus (GEO) and Qilu Hospital of Shandong University. We further assessed the immune microenvironment of patients with high and low CDI scores. Moreover, the expression of the signature genes in HNSCC cell lines were explored.
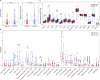
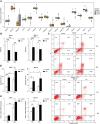
Results: We found that CDI was an independent prognostic indicator for overall survival (hazard ratio 3.80, 95% confidential interval: 2.70-5.40, P < 0.001). Furthermore, HNSCC patients with high CDI scores obtained increased overall survival post radiation indicating benefits from radiotherapy of this subgroup. On the other hand, HPV negative HNSCC patients with low CDI exhibited increased checkpoint gene expressions, an inflamed tumor microenvironment and an enriched immune response-related functions, suggesting the potential benefits from checkpoint immunotherapies of this subgroup. Moreover, we validated the baseline and induced expressions of above 16 genes in two HPV negative HNSCC cell lines, CAL27 and SCC-15.
Discussion: We established a prognostic signature and emphasized its implements in the therapeutic choices of HPV negative HNSCC patients, the majority and the poor outcome population of HNSCC.
Keywords: apoptosis; autophagy; head and neck squamous cell carcinoma; human papillomavirus; prognosis; pyroptosis; radiotherapy; tumor microenvironment.
Copyright © 2023 Nan, Dou, Chen, Wang, Sun, Meng, Neckenig, Ai, Liu, Dong, Ma, Cheng and Qu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of pyroptosis-related gene prognostic signature in head and neck squamous cell carcinoma.Cancer Med. 2022 Dec;11(24):5129-5144. doi: 10.1002/cam4.4825. Epub 2022 May 16. Cancer Med. 2022. PMID: 35574984 Free PMC article.
-
A novel Pyroptosis-related long non-coding RNA signature for predicting the prognosis and immune landscape of head and neck squamous cell carcinoma.Cancer Med. 2022 Dec;11(24):5097-5112. doi: 10.1002/cam4.4819. Epub 2022 May 14. Cancer Med. 2022. PMID: 35567376 Free PMC article. Clinical Trial.
-
A pyroptosis-related gene expression signature predicts immune microenvironment and prognosis in head and neck squamous cell carcinoma.Eur Arch Otorhinolaryngol. 2024 Feb;281(2):953-963. doi: 10.1007/s00405-023-08316-y. Epub 2023 Dec 8. Eur Arch Otorhinolaryngol. 2024. PMID: 38063904
-
[Mutation signatures in head and neck squamous cell carcinoma : Pathogenesis and therapeutic potential].HNO. 2020 Dec;68(12):922-926. doi: 10.1007/s00106-020-00954-6. Epub 2020 Oct 12. HNO. 2020. PMID: 33044581 Review. German.
-
Immune microenvironment of human papillomavirus-positive head and neck squamous cell carcinoma.Sci Prog. 2024 Jan-Mar;107(1):368504241237888. doi: 10.1177/00368504241237888. Sci Prog. 2024. PMID: 38545800 Free PMC article. Review.
Cited by
-
IL-37 attenuated HPV induced inflammation and growth of oral epithelial cells via regulating autophagy.Heliyon. 2024 Jul 24;10(15):e35131. doi: 10.1016/j.heliyon.2024.e35131. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39157375 Free PMC article.
-
The role of novel programmed cell death in head and neck squamous cell carcinoma: from mechanisms to potential therapies.Front Pharmacol. 2023 Sep 25;14:1228985. doi: 10.3389/fphar.2023.1228985. eCollection 2023. Front Pharmacol. 2023. PMID: 37818196 Free PMC article. Review.
-
Exploring Autophagy-Related Gene Expression in Hepatocellular Carcinoma via TCGA, GEPIA2, and HPA Databases: Implications for Prognosis.J Hepatocell Carcinoma. 2025 Jul 22;12:1557-1586. doi: 10.2147/JHC.S520917. eCollection 2025. J Hepatocell Carcinoma. 2025. PMID: 40726620 Free PMC article.
-
Leveraging cell death patterns to predict metastasis in prostate adenocarcinoma and targeting PTGDS for tumor suppression.Sci Rep. 2024 Sep 17;14(1):21680. doi: 10.1038/s41598-024-72985-w. Sci Rep. 2024. PMID: 39289451 Free PMC article.
-
Elucidating the molecular and immune interplay between head and neck squamous cell carcinoma and diffuse large B-cell lymphoma through bioinformatics and machine learning.Transl Cancer Res. 2024 Nov 30;13(11):5725-5750. doi: 10.21037/tcr-24-1064. Epub 2024 Nov 21. Transl Cancer Res. 2024. PMID: 39697749 Free PMC article.
References
-
- Lacas B, Carmel A, Landais C, Wong SJ, Licitra L, Tobias JS, et al. . Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): An update on 107 randomized trials and 19,805 patients, on behalf of MACH-NC group. Radiother Oncol (2021) 156:281–93. doi: 10.1016/j.radonc.2021.01.013 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

