Establishment and maintenance of DNA methylation in nematode feeding sites
- PMID: 36704169
- PMCID: PMC9873351
- DOI: 10.3389/fpls.2022.1111623
Establishment and maintenance of DNA methylation in nematode feeding sites
Abstract
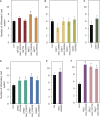
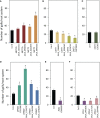
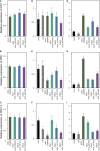
A growing body of evidence indicates that epigenetic mechanisms, particularly DNA methylation, play key regulatory roles in plant-nematode interactions. Nevertheless, the transcriptional activity of key genes mediating DNA methylation and active demethylation in the nematode feeding sites remains largely unknown. Here, we profiled the promoter activity of 12 genes involved in maintenance and de novo establishment of DNA methylation and active demethylation in the syncytia and galls induced respectively by the cyst nematode Heterodera schachtii and the root-knot nematode Meloidogyne incognita in Arabidopsis roots. The promoter activity assays revealed that expression of the CG-context methyltransferases is restricted to feeding site formation and development stages. Chromomethylase1 (CMT1), CMT2, and CMT3 and Domains Rearranged Methyltransferase2 (DRM2) and DRM3, which mediate non-CG methylation, showed similar and distinct expression patterns in the syncytia and galls at various time points. Notably, the promoters of various DNA demethylases were more active in galls as compared with the syncytia, particularly during the early stage of infection. Mutants impaired in CG or CHH methylation similarly enhanced plant susceptibility to H. schachtii and M. incognita, whereas mutants impaired in CHG methylation reduced plant susceptibility only to M. incognita. Interestingly, hypermethylated mutants defective in active DNA demethylation exhibited contrasting responses to infection by H. schachtii and M. incognita, a finding most likely associated with differential regulation of defense-related genes in these mutants upon nematode infection. Our results point to methylation-dependent mechanisms regulating plant responses to infection by cyst and root-knot nematodes.
Keywords: Arabidopsis; DNA methylation; Heterodera schachtii; Meloidogyne incognita; active DNA demethylation; gall; promoter activity; syncytium.
Copyright © 2023 Bennett, Hawk, Lopes-Caitar, Adams, Rice and Hewezi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The DNA methylation landscape of the root-knot nematode-induced pseudo-organ, the gall, in Arabidopsis, is dynamic, contrasting over time, and critically important for successful parasitism.New Phytol. 2022 Dec;236(5):1888-1907. doi: 10.1111/nph.18395. Epub 2022 Sep 2. New Phytol. 2022. PMID: 35872574 Free PMC article.
-
First report of Meloidogyne javanica on Ginger and Turmeric in the United States.J Nematol. 2019;51:1-3. doi: 10.21307/jofnem-2019-006. J Nematol. 2019. PMID: 31088018 Free PMC article.
-
Image-Based Lignin Detection in Nematode-Induced Feeding Sites in Arabidopsis Roots.Bio Protoc. 2025 May 5;15(9):e5301. doi: 10.21769/BioProtoc.5301. eCollection 2025 May 5. Bio Protoc. 2025. PMID: 40364986 Free PMC article.
-
Modulation of Arabidopsis Flavonol Biosynthesis Genes by Cyst and Root-Knot Nematodes.Plants (Basel). 2020 Feb 17;9(2):253. doi: 10.3390/plants9020253. Plants (Basel). 2020. PMID: 32079157 Free PMC article.
-
Differential gene expression in Arabidopsis following infection by plant-parasitic nematodes Meloidogyne incognita and Heterodera schachtii.Mol Plant Pathol. 2007 Sep;8(5):595-609. doi: 10.1111/j.1364-3703.2007.00416.x. Mol Plant Pathol. 2007. PMID: 20507524
Cited by
-
A Critical Appraisal of DNA Transfer from Plants to Parasitic Cyst Nematodes.Mol Biol Evol. 2024 Feb 1;41(2):msae030. doi: 10.1093/molbev/msae030. Mol Biol Evol. 2024. PMID: 38366574 Free PMC article.
-
The other side of the coin: systemic effects of Serendipita indica root colonization on development of sedentary plant-parasitic nematodes in Arabidopsis thaliana.Planta. 2024 Apr 14;259(5):121. doi: 10.1007/s00425-024-04402-5. Planta. 2024. PMID: 38615288 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

