Rapid detection of SARS-CoV-2: The gradual boom of lateral flow immunoassay
- PMID: 36704307
- PMCID: PMC9871317
- DOI: 10.3389/fbioe.2022.1090281
Rapid detection of SARS-CoV-2: The gradual boom of lateral flow immunoassay
Abstract
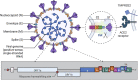
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is still in an epidemic situation, which poses a serious threat to the safety of people and property. Rapid diagnosis and isolation of infected individuals are one of the important methods to control virus transmission. Existing lateral flow immunoassay techniques have the advantages of rapid, sensitive, and easy operation, and some new options have emerged with the continuous development of nanotechnology. Such as lateral flow immunoassay test strips based on colorimetric-fluorescent dual-mode and gold nanoparticles, Surface Enhanced Raman Scattering, etc., these technologies have played an important role in the rapid diagnosis of COVID-19. In this paper, we summarize the current research progress of lateral flow immunoassay in the field of Severe Acute Respiratory Syndrome Coronavirus 2 infection diagnosis, analyze the performance of Severe Acute Respiratory Syndrome Coronavirus 2 lateral flow immunoassay products, review the advantages and limitations of different detection methods and markers, and then explore the competitive CRISPR-based nucleic acid chromatography detection method. This method combines the advantages of gene editing and lateral flow immunoassay and can achieve rapid and highly sensitive lateral flow immunoassay detection of target nucleic acids, which is expected to be the most representative method for community and clinical point-of-care testing. We hope that researchers will be inspired by this review and strive to solve the problems in the design of highly sensitive targets, the selection of detection methods, and the enhancement of CRISPR technology, to truly achieve rapid, sensitive, convenient, and specific detection of novel coronaviruses, thus promoting the development of novel coronavirus diagnosis and contributing our modest contribution to the world's fight against epidemics.
Keywords: COVID-19; CRISPR; SARS-CoV-2; antibody; antigen; lateral flow immunoassay; nanotechnology; nucleic acid.
Copyright © 2023 He, Zhu, Zhou, Jiang, Yin, Su, Zhang, Chen and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Next-Generation Rapid and Ultrasensitive Lateral Flow Immunoassay for Detection of SARS-CoV-2 Variants.ACS Sens. 2023 Oct 27;8(10):3733-3743. doi: 10.1021/acssensors.3c01019. Epub 2023 Sep 7. ACS Sens. 2023. PMID: 37675933 Clinical Trial.
-
[Applications of separation technology in novel coronavirus research, epidemic prevention and detection].Se Pu. 2021 Jul 8;39(7):679-685. doi: 10.3724/SP.J.1123.2021.03022. Se Pu. 2021. PMID: 34227364 Free PMC article. Review. Chinese.
-
Development and Efficacy of Lateral Flow Point-of-Care Testing Devices for Rapid and Mass COVID-19 Diagnosis by the Detections of SARS-CoV-2 Antigen and Anti-SARS-CoV-2 Antibodies.Diagnostics (Basel). 2021 Sep 24;11(10):1760. doi: 10.3390/diagnostics11101760. Diagnostics (Basel). 2021. PMID: 34679458 Free PMC article. Review.
-
Simultaneous Detection of SARS-CoV-2 IgG/IgM Antibodies, Using Gold Nanoparticles-Based Lateral Flow Immunoassay.Monoclon Antib Immunodiagn Immunother. 2021 Oct;40(5):210-218. doi: 10.1089/mab.2021.0027. Monoclon Antib Immunodiagn Immunother. 2021. PMID: 34678096
-
Optical lateral flow assays in early diagnosis of SARS-CoV-2 infection.Anal Sci. 2024 Sep;40(9):1571-1591. doi: 10.1007/s44211-024-00596-6. Epub 2024 May 17. Anal Sci. 2024. PMID: 38758251 Review.
Cited by
-
BRET-based biosensors for SARS-CoV-2 oligonucleotide detection.Front Bioeng Biotechnol. 2024 Jun 3;12:1353479. doi: 10.3389/fbioe.2024.1353479. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38887615 Free PMC article.
-
Comparison of Three Lateral Flow Immunoassay Formats for the Detection of Antibodies against the SARS-CoV-2 Antigen.Biosensors (Basel). 2023 Jul 20;13(7):750. doi: 10.3390/bios13070750. Biosensors (Basel). 2023. PMID: 37504148 Free PMC article.
-
Advancements in the synergy of isothermal amplification and CRISPR-cas technologies for pathogen detection.Front Bioeng Biotechnol. 2023 Oct 10;11:1273988. doi: 10.3389/fbioe.2023.1273988. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37885449 Free PMC article. Review.
-
Organokines in COVID-19: A Systematic Review.Cells. 2023 May 9;12(10):1349. doi: 10.3390/cells12101349. Cells. 2023. PMID: 37408184 Free PMC article.
-
Recent Studies on Metal-Embedded Silica Nanoparticles for Biological Applications.Nanomaterials (Basel). 2024 Jan 26;14(3):268. doi: 10.3390/nano14030268. Nanomaterials (Basel). 2024. PMID: 38334538 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

